Abstract
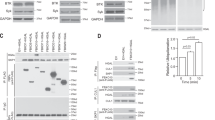
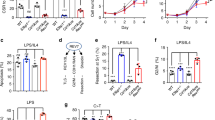
The enzyme activation-induced deaminase (AID) triggers antibody diversification in B cells by catalyzing deamination and consequently mutation of immunoglobulin genes. To minimize off-target deamination, AID is restrained by several regulatory mechanisms including nuclear exclusion, thought to be mediated exclusively by active nuclear export. Here we identify two other mechanisms involved in controlling AID subcellular localization. AID is unable to passively diffuse into the nucleus, despite its small size, and its nuclear entry requires active import mediated by a conformational nuclear localization signal. We also identify in its C terminus a determinant for AID cytoplasmic retention, which hampers diffusion to the nucleus, competes with nuclear import and is crucial for maintaining the predominantly cytoplasmic localization of AID in steady-state conditions. Blocking nuclear import alters the balance between these processes in favor of cytoplasmic retention, resulting in reduced isotype class switching.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Muramatsu, M. et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 (2000).
Revy, P. et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102, 565–575 (2000).
Peled, J.U. et al. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 26, 481–511 (2007).
Di Noia, J.M. & Neuberger, M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76, 1–22 (2007).
Chaudhuri, J. et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94, 157–214 (2007).
Martin, A. & Scharff, M.D. Somatic hypermutation of the AID transgene in B and non-B cells. Proc. Natl. Acad. Sci. USA 99, 12304–12308 (2002).
Okazaki, I.M. et al. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197, 1173–1181 (2003).
Liu, M. et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature 451, 841–845 (2008).
Pasqualucci, L. et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. USA 95, 11816–11821 (1998).
Shen, H.M., Peters, A., Baron, B., Zhu, X. & Storb, U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 280, 1750–1752 (1998).
Dorsett, Y. et al. A role for AID in chromosome translocations between c-myc and the IgH variable region. J. Exp. Med. 204, 2225–2232 (2007).
Ramiro, A.R. et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118, 431–438 (2004).
Crouch, E.E. et al. Regulation of AID expression in the immune response. J. Exp. Med. 204, 1145–1156 (2007).
de Yébenes, V.G. et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 205, 2199–2206 (2008).
Dorsett, Y. et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity 28, 630–638 (2008).
Teng, G. et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 28, 621–629 (2008).
Ito, S. et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. USA 101, 1975–1980 (2004).
McBride, K.M., Barreto, V.M., Ramiro, A.R., Stavropoulos, P. & Nussenzweig, M.C. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199, 1235–1244 (2004).
Aoufouchi, S. et al. Proteasomal degradation restricts the nuclear lifespan of AID. J. Exp. Med. 205, 1357–1368 (2008).
Basu, U. et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature 438, 508–511 (2005).
McBride, K.M. et al. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc. Natl. Acad. Sci. USA 103, 8798–8803 (2006).
Pasqualucci, L., Kitaura, Y., Gu, H. & Dalla-Favera, R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc. Natl. Acad. Sci. USA 103, 395–400 (2006).
Rada, C., Jarvis, J.M. & Milstein, C. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc. Natl. Acad. Sci. USA 99, 7003–7008 (2002).
Brar, S.S., Watson, M. & Diaz, M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 279, 26395–26401 (2004).
Richardson, W.D., Mills, A.D., Dilworth, S.M., Laskey, R.A. & Dingwall, C. Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell 52, 655–664 (1988).
Newmeyer, D.D. & Forbes, D.J. Nuclear import can be separated into distinct steps in vitro: nuclear pore binding and translocation. Cell 52, 641–653 (1988).
Guiochon-Mantel, A. et al. Nucleocytoplasmic shuttling of the progesterone receptor. EMBO J. 10, 3851–3859 (1991).
Larijani, M. et al. AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequence-independent manner. Mol. Cell. Biol. 27, 20–30 (2007).
Macara, I.G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65, 570–594 (2001).
Görlich, D. & Kutay, U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660 (1999).
Chen, K.M. et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452, 116–119 (2008).
Holden, L.G. et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 456, 121–124 (2008).
Prochnow, C. et al. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature 445, 447–451 (2007).
Nilsen, H. et al. Analysis of uracil-DNA glycosylases from the murine Ung gene reveals differential expression in tissues and in embryonic development and a subcellular sorting pattern that differs from the human homologues. Nucleic Acids Res. 28, 2277–2285 (2000).
Conticello, S.G. et al. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol. Cell 31, 474–484 (2008).
Lange, A. et al. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282, 5101–5105 (2007).
Miyamoto, Y. et al. Cellular stresses induce the nuclear accumulation of importin α and cause a conventional nuclear import block. J. Cell Biol. 165, 617–623 (2004).
Kodiha, M., Chu, A., Matusiewicz, N. & Stochaj, U. Multiple mechanisms promote the inhibition of classical nuclear import upon exposure to severe oxidative stress. Cell Death Differ. 11, 862–874 (2004).
Shivarov, V., Shinkura, R. & Honjo, T. Dissociation of in vitro DNA deamination activity and physiological functions of AID mutants. Proc. Natl. Acad. Sci. USA 105, 15866–15871 (2008).
Doi, T. et al. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc. Natl. Acad. Sci. USA 106, 2758–2763 (2009).
Barreto, V., Reina-San-Martin, B., Ramiro, A.R., McBride, K.M. & Nussenzweig, M.C. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell 12, 501–508 (2003).
Ta, V. et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat. Immunol. 4, 843–848 (2003).
Shinkura, R. et al. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 5, 707–712 (2004).
LaCasse, E.C. & Lefebvre, Y.A. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 23, 1647–1656 (1995).
Chester, A. et al. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J. 22, 3971–3982 (2003).
Yang, Y. & Smith, H.C. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc. Natl. Acad. Sci. USA 94, 13075–13080 (1997).
Dickerson, S.K., Market, E., Besmer, E. & Papavasiliou, F.N. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197, 1291–1296 (2003).
Yang, G. et al. Activation-induced deaminase cloning, localization, and protein extraction from young VH-mutant rabbit appendix. Proc. Natl. Acad. Sci. USA 102, 17083–17088 (2005).
Bennett, R.P., Presnyak, V., Wedekind, J.E. & Smith, H.C. Nuclear exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J. Biol. Chem. 283, 7320–7327 (2008).
Lau, P.P., Zhu, H.J., Baldini, A., Charnsangavej, C. & Chan, L. Dimeric structure of a human apolipoprotein B mRNA editing protein and cloning and chromosomal localization of its gene. Proc. Natl. Acad. Sci. USA 91, 8522–8526 (1994).
Stenglein, M.D., Matsuo, H. & Harris, R.S. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J. Virol. 82, 9591–9599 (2008).
Wu, X., Geraldes, P., Platt, J.L. & Cascalho, M. The double-edged sword of activation-induced cytidine deaminase. J. Immunol. 174, 934–941 (2005).
Cattoretti, G. et al. Nuclear and cytoplasmic AID in extrafollicular and germinal center B cells. Blood 107, 3967–3975 (2006).
Greiner, A. et al. Differential expression of activation-induced cytidine deaminase (AID) in nodular lymphocyte-predominant and classical Hodgkin lymphoma. J. Pathol. 205, 541–547 (2005).
Pasqualucci, L. et al. Expression of the AID protein in normal and neoplastic B cells. Blood 104, 3318–3325 (2004).
Petersen-Mahrt, S.K., Harris, R.S. & Neuberger, M.S. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418, 99–104 (2002).
Di Noia, J.M. et al. Dependence of antibody gene diversification on uracil excision. J. Exp. Med. 204, 3209–3219 (2007).
Kitamura, T. et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31, 1007–1014 (2003).
Zhang, W. et al. Clonal instability of V region hypermutation in the Ramos Burkitt's lymphoma cell line. Int. Immunol. 13, 1175–1184 (2001).
Acknowledgements
We thank S. Conticello and M. Neuberger (Medical Research Council Laboratory of Molecular Biology), I. Macara (University of Virginia School of Medicine), M. Malim (King's College London School of Medicine), N. Navaratnam (Imperial College London), A. Martin (Univeristy of Toronto), S. Swaminathan (University of Florida), J. Archambault, E. Cohen, T. Möröy and Y. Guindon (Institut de Recherches Cliniques de Montréal (IRCM)) for providing reagents and T. Honjo (Kyoto University School of Medicine) for the AID-deficient mice through A. Lamarre (INRS-Institute Armand-Frappier). We thank E.-L. Thivierge and C. Toulouse for expert animal care and D. Filion for help with confocal microscopy. We are grateful to H. Krokan and R. Harris for discussions and to S. Conticello, C. Buscaglia, D. Muñoz and J.F. Côté for critically reading the manuscript. This work was supported by the Canadian Institutes of Health Research (MOP 84543) and a Canada Research Chair (to J.M.D.). A.O. was supported by a fellowship from the Canadian Institutes of Health Research Cancer Training Program at the IRCM. V.A.C. was supported in part by a Michel Saucier fellowship from the Louis-Pasteur Canadian Fund through the University of Montreal.
Author information
Authors and Affiliations
Contributions
A.-M.P., A.O., Y.H., V.A.C., A.B. and J.M.D. performed research; all authors analyzed data and discussed results; J.M.D. designed research and wrote the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Table 1 and Supplementary Methods (PDF 4516 kb)
Rights and permissions
About this article
Cite this article
Patenaude, AM., Orthwein, A., Hu, Y. et al. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol 16, 517–527 (2009). https://doi.org/10.1038/nsmb.1598
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1598
This article is cited by
-
USP10 regulates B cell response to SARS-CoV-2 or HIV-1 nanoparticle vaccines through deubiquitinating AID
Signal Transduction and Targeted Therapy (2022)
-
Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination
Nature Reviews Genetics (2022)
-
Plasma cells, plasmablasts, and AID+/CD30+ B lymphoblasts inside and outside germinal centres: details of the basal light zone and the outer zone in human palatine tonsils
Histochemistry and Cell Biology (2020)
-
A licensing step links AID to transcription elongation for mutagenesis in B cells
Nature Communications (2018)
-
Diversity of Immunoglobulin (Ig) Isotypes and the Role of Activation-Induced Cytidine Deaminase (AID) in Fish
Molecular Biotechnology (2018)