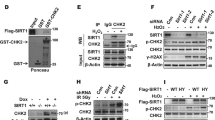
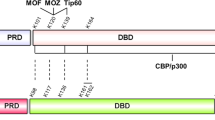
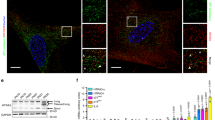
Abstract
Cellular senescence is one of the key strategies to suppress expansion of cells with mutations. Senescence is induced in response to genotoxic and oxidative stress. Here we show that the transcription factor Bach1 (BTB and CNC homology 1, basic leucine zipper transcription factor 1), which inhibits oxidative stress-inducible genes, is a crucial negative regulator of oxidative stress–induced cellular senescence. Bach1-deficient murine embryonic fibroblasts showed a propensity to undergo more rapid and profound p53-dependent premature senescence than control wild-type cells in response to oxidative stress. Bach1 formed a complex that contained p53, histone deacetylase 1 and nuclear co-repressor N-coR. Bach1 was recruited to a subset of p53 target genes and contributed to impeding p53 action by promoting histone deacetylation. Because Bach1 is regulated by oxidative stress and heme, our data show that Bach1 connects oxygen metabolism and cellular senescence as a negative regulator of p53.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Braig, M. et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 (2005).
Chen, Z. et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730 (2005).
Michaloglou, C. et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 (2005).
Collado, M. et al. Tumour biology: senescence in premalignant tumours. Nature 436, 642 (2005).
Ben-Porath, I. & Weinberg, R.A. The signals and pathways activating cellular senescence. Int. J. Biochem. Cell Biol. 37, 961–976 (2005).
Brown, J.P., Wei, W. & Sedivy, J.M. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 277, 831–834 (1997).
Kortlever, R.M., Higgins, P.J. & Bernards, R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat. Cell Biol. 8, 878–884 (2006).
Kubbutat, M.H., Jones, S.N. & Vousden, K.H. Regulation of p53 stability by MDM2. Nature 387, 299–303 (1997).
Honda, R., Tanaka, H. & Yasuda, H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 (1997).
Shieh, S.Y., Ikeda, M., Taya, Y. & Prives, C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91, 325–334 (1997).
Finkel, T. Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 15, 247–254 (2003).
Parrinello, S. et al. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5, 741–747 (2003).
Harvey, M. et al. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene 8, 2457–2467 (1993).
Oyake, T. et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16, 6083–6095 (1996).
Igarashi, K. & Sun, J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid. Redox Signal. 8, 107–118 (2006).
Sun, J. et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 21, 5216–5224 (2002).
Omura, S. et al. Effects of genetic ablation of bach1 upon smooth muscle cell proliferation and atherosclerosis after cuff injury. Genes Cells 10, 277–285 (2005).
Dimri, G.P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92, 9363–9367 (1995).
Rogakou, E.P., Pilch, D.R., Orr, A.H., Ivanova, V.S. & Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Lowe, S.W., Cepero, E. & Evan, G. Intrinsic tumour suppression. Nature 432, 307–315 (2004).
Gire, V. & Wynford-Thomas, D. Reinitiation of DNA synthesis and cell division in senescent human fibroblasts by microinjection of anti-p53 antibodies. Mol. Cell. Biol. 18, 1611–1621 (1998).
Campisi, J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120, 513–522 (2005).
Serrano, M., Lin, A.W., McCurrach, M.E., Beach, D. & Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Attardi, L.D. et al. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 14, 704–718 (2000).
Clarke, A.R. et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362, 849–852 (1993).
Wei, C.L. et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219 (2006).
Tanaka, H. et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404, 42–49 (2000).
Zoidl, G. et al. Influence of elevated expression of rat wild-type PMP22 and its mutant PMP22Trembler on cell growth of NIH3T3 fibroblasts. Cell Tissue Res. 287, 459–470 (1997).
Gu, W. & Roeder, R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 (1997).
Ikura, T. et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 (2000).
Fujiwara, K.T., Kataoka, K. & Nishizawa, M. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene 8, 2371–2380 (1993).
Yamasaki, C., Tashiro, S., Nishito, Y., Sueda, T. & Igarashi, K. Dynamic cytoplasmic anchoring of the transcription factor Bach1 by intracellular hyaluronic acid binding protein IHABP. J. Biochem. 137, 287–296 (2005).
Heinzel, T. et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387, 43–48 (1997).
Alland, L. et al. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387, 49–55 (1997).
Reczek, E.E., Flores, E.R., Tsay, A.S., Attardi, L.D. & Jacks, T. Multiple response elements and differential p53 binding control Perp expression during apoptosis. Mol. Cancer Res. 1, 1048–1057 (2003).
Kaeser, M.D. & Iggo, R.D. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl. Acad. Sci. USA 99, 95–100 (2002).
Sun, J. et al. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc. Natl. Acad. Sci. USA 101, 1461–1466 (2004).
de Stanchina, E. et al. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell 13, 523–535 (2004).
Pearson, M. & Pelicci, P.G. PML interaction with p53 and its role in apoptosis and replicative senescence. Oncogene 20, 7250–7256 (2001).
Yang, A. et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305–316 (1998).
Yang, A. et al. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell 24, 593–602 (2006).
Hublitz, P. et al. NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes Dev. 19, 2912–2924 (2005).
Sablina, A.A. et al. The antioxidant function of the p53 tumor suppressor. Nat. Med. 11, 1306–1313 (2005).
Ishikawa, M., Numazawa, S. & Yoshida, T. Redox regulation of the transcriptional repressor Bach1. Free Radic. Biol. Med. 38, 1344–1352 (2005).
Igarashi, K. et al. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367, 568–572 (1994).
Igarashi, K. et al. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for β-globin locus control region complex. J. Biol. Chem. 273, 11783–11790 (1998).
Ito, A. et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20, 1331–1340 (2001).
Acknowledgements
We thank T. Jacks (Massachusetts Institute of Technology), T.P. Yao (Duke University), K. Tanimoto (Hiroshima University), H. Ogawa (National Institute of Basic Biology), N. Tanaka (Nippon Medical School) and K. Umesono for providing plasmids and antibodies; Y. Ishikawa and K. Nakayama (Tohoku University) for providing p53−/− MEFs; and M. Katsuki (National Institute of Basic Biology) for providing Trp53−/− mice. We also thank M. Kobayashi and Y. Taya for comments on the manuscript; S. Tashiro and T. Ide for valuable advice to initiate the project; M. Ikura for help in cell culturing; and M. Yoshizumi and N. Tanaka for advice. This work was supported by Grants-in-aid and the Network Medicine Global-COE Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and grants from the Uehara Foundation, the Takeda Foundation and the Astellas Foundation for Research on Metabolic Disorders.
Author information
Authors and Affiliations
Contributions
Y.D., T.I., Y.K., K.O., A.N., A.M., S.O., A.I. and M.Y. contributed to performing and assisting with experiments; A.M. and T.O. contributed to performing gene expression profiling; Y.H. and T.N. contributed to performing MS/MS analysis; K.I. designed and conceptualized the study; Y.D., T.I., T.N. and K.I. interpreted the data and Y.D., T.I. and K.I. wrote the manuscript. All authors made comments on the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Methods (PDF 243 kb)
Rights and permissions
About this article
Cite this article
Dohi, Y., Ikura, T., Hoshikawa, Y. et al. Bach1 inhibits oxidative stress–induced cellular senescence by impeding p53 function on chromatin. Nat Struct Mol Biol 15, 1246–1254 (2008). https://doi.org/10.1038/nsmb.1516
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1516
This article is cited by
-
Specific regulation of BACH1 by the hotspot mutant p53R175H reveals a distinct gain-of-function mechanism
Nature Cancer (2023)
-
The dual role and mutual dependence of heme/HO-1/Bach1 axis in the carcinogenic and anti-carcinogenic intersection
Journal of Cancer Research and Clinical Oncology (2023)
-
Comparative parallel multi-omics analysis during the induction of pluripotent and trophectoderm states
Nature Communications (2022)
-
Lipid peroxidation and the subsequent cell death transmitting from ferroptotic cells to neighboring cells
Cell Death & Disease (2021)
-
Regulatory mechanisms of heme regulatory protein BACH1: a potential therapeutic target for cancer
Medical Oncology (2021)