Abstract
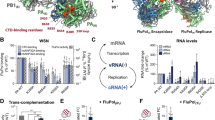
The replication of many retroviruses is mediated by a transcriptional activator protein, Tat, which activates RNA polymerase II at the level of transcription elongation. Tat interacts with Cyclin T1 of the positive transcription-elongation factor P-TEFb to recruit the transactivation-response TAR RNA, which acts as a promoter element in the transcribed 5′ end of the viral long terminal repeat. Here we present the structure of the cyclin box domain of Cyclin T1 in complex with the Tat protein from the equine infectious anemia virus and its corresponding TAR RNA. The basic RNA-recognition motif of Tat adopts a helical structure whose flanking regions interact with a cyclin T–specific loop in the first cyclin box repeat. Together, both proteins coordinate the stem-loop structure of TAR. Our findings show that Tat binds to a surface on Cyclin T1 similar to where recognition motifs from substrate and inhibitor peptides were previously found to interact within Cdk–cyclin pairs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Stevenson, M. HIV-1 pathogenesis. Nat. Med. 9, 853–860 (2003).
Jones, K.A. & Peterlin, B.M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63, 717–743 (1994).
Karn, J. Tackling Tat. J. Mol. Biol. 293, 235–254 (1999).
Barboric, M. & Peterlin, B.M. A new paradigm in eukaryotic biology: HIV Tat and the control of transcriptional elongation. PLoS Biol. 3, e76 (2005).
Rosen, C.A., Sodroski, J.G. & Haseltine, W.A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell 41, 813–823 (1985).
Okamoto, T. & Wong-Staal, F. Demonstration of virus-specific transcriptional activator(s) in cells infected with HTLV-III by an in vitro cell-free system. Cell 47, 29–35 (1986).
Sodroski, J. et al. Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science 227, 171–173 (1985).
Wei, P., Garber, M.E., Fang, S.M., Fischer, W.H. & Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92, 451–462 (1998).
Saunders, A., Core, L.J. & Lis, J.T. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7, 557–567 (2006).
Sims, R.J., III, Belotserkovskaya, R. & Reinberg, D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18, 2437–2468 (2004).
Shilatifard, A., Conaway, R.C. & Conaway, J.W. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 72, 693–715 (2003).
Meinhart, A., Kamenski, T., Hoeppner, S., Baumli, S. & Cramer, P. A structural perspective of CTD function. Genes Dev. 19, 1401–1415 (2005).
Ni, Z. et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell. Biol. 28, 1161–1170 (2008).
Garriga, J. et al. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol. Cell. Biol. 23, 5165–5173 (2003).
Yang, Z., Zhu, Q., Luo, K. & Zhou, Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414, 317–322 (2001).
Nguyen, V.T., Kiss, T., Michels, A.A. & Bensaude, O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414, 322–325 (2001).
Michels, A.A. et al. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 23, 4859–4869 (2003).
Yik, J.H. et al. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12, 971–982 (2003).
Schulte, A. et al. Identification of a Cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 280, 24968–24977 (2005).
Barboric, M. et al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35, 2003–2012 (2007).
Taube, R., Fujinaga, K., Wimmer, J., Barboric, M. & Peterlin, B.M. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264, 245–253 (1999).
Garber, M.E. et al. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12, 3512–3527 (1998).
Leroux, C., Cadoré, J.L. & Montelaro, R.C. Equine infectious anemia virus (EIAV): what has HIV's country cousin got to tell us? Vet. Res. 35, 485–512 (2004).
Fujinaga, K. et al. A minimal chimera of human cyclin T1 and Tat binds TAR and activates human immunodeficiency virus transcription in murine cells. J. Virol. 76, 12934–12939 (2002).
Anand, K., Schulte, A., Fujinaga, K., Scheffzek, K. & Geyer, M. Cyclin box structure of the P-TEFb subunit Cyclin T1 derived from a fusion complex with EIAV Tat. J. Mol. Biol. 370, 826–836 (2007).
Noble, M.E., Endicott, J.A., Brown, N.R. & Johnson, L.N. The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends Biochem. Sci. 22, 482–487 (1997).
Derse, D. & Newbold, S.H. Mutagenesis of EIAV TAT reveals structural features essential for transcriptional activation and TAR element recognition. Virology 194, 530–536 (1993).
Lee, J.C. & Gutell, R.R. Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. J. Mol. Biol. 344, 1225–1249 (2004).
Hetzer, C., Dormeyer, W., Schnölzer, M. & Ott, M. Decoding Tat: the biology of HIV Tat posttranslational modifications. Microbes Infect. 7, 1364–1369 (2005).
Bieniasz, P.D., Grdina, T.A., Bogerd, H.P. & Cullen, B.R. Highly divergent lentiviral Tat proteins activate viral gene expression by a common mechanism. Mol. Cell. Biol. 19, 4592–4599 (1999).
Taube, R. et al. Interactions between equine Cyclin T1, Tat, and TAR are disrupted by a leucine-to-valine substitution found in human Cyclin T1. J. Virol. 74, 892–898 (2000).
Richter, S., Cao, H. & Rana, T.M. Specific HIV-1 TAR RNA loop sequence and functional groups are required for human Cyclin T1-Tat-TAR ternary complex formation. Biochemistry 41, 6391–6397 (2002).
Puglisi, J.D., Chen, L., Blanchard, S. & Frankel, A.D. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science 270, 1200–1203 (1995).
Ye, X., Kumar, R.A. & Patel, D.J. Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex. Chem. Biol. 2, 827–840 (1995).
Calabro, V., Daugherty, M.D. & Frankel, A.D. A single intermolecular contact mediates intramolecular stabilization of both RNA and protein. Proc. Natl. Acad. Sci. USA 102, 6849–6854 (2005).
Howl, J., Nicholl, I.D. & Jones, S. The many futures for cell-penetrating peptides: how soon is now? Biochem. Soc. Trans. 35, 767–769 (2007).
Richter, S., Ping, Y.H. & Rana, T.M. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA 99, 7928–7933 (2002).
Aboul-ela, F., Karn, J. & Varani, G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 253, 313–332 (1995).
Ippolito, J.A. & Steitz, T.A. A 1.3- resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc. Natl. Acad. Sci. USA 95, 9819–9824 (1998).
Brown, N.R., Noble, M.E., Endicott, J.A. & Johnson, L.N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1, 438–443 (1999).
Cheng, K.Y. et al. The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition. J. Biol. Chem. 281, 23167–23179 (2006).
Russo, A.A., Jeffrey, P.D., Patten, A.K., Massague, J. & Pavletich, N.P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382, 325–331 (1996).
Dames, S.A. et al. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc. Natl. Acad. Sci. USA 104, 14312–14317 (2007).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C. & Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. 61, 458–464 (2005).
Emsley, P. & Cowtan, K. COOT: model-building tools for molecular graphics. Acta Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr 58, 1948–1954 (2002).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Acknowledgements
We thank D. Ludwig for expert technical assistance and P. Afonine for helpful discussions on the PHENIX program. Data collection was done at the European Synchrotron Radiation Facility, beamline ID23-1, Grenoble, France, and we thank the beamline staff for assistance. M.G. thanks R. S. Goody for discussions and continuous support. K.A. was funded previously by the European Molecular Biology Organization and at present by a Marie-Curie fellowship. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to M.G. (GE 976/5).
Author information
Authors and Affiliations
Contributions
K.A. crystallized the CycT1–Tat–TAR complex and solved the structure; A.S. purified the proteins and performed biochemical characterizations; K.V.B. made RNA transcripts and gel shift assays; K.S. and M.G. assisted K.A. throughout structure determination; M.G. supervised the study, designed all constructs and wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 845 kb)
Rights and permissions
About this article
Cite this article
Anand, K., Schulte, A., Vogel-Bachmayr, K. et al. Structural insights into the Cyclin T1–Tat–TAR RNA transcription activation complex from EIAV. Nat Struct Mol Biol 15, 1287–1292 (2008). https://doi.org/10.1038/nsmb.1513
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1513
This article is cited by
-
Differences in Transcriptional Dynamics Between T-cells and Macrophages as Determined by a Three-State Mathematical Model
Scientific Reports (2020)
-
Herb-target interaction network analysis helps to disclose molecular mechanism of traditional Chinese medicine
Scientific Reports (2016)
-
A single point mutation in cyclin T1 eliminates binding to Hexim1, Cdk9 and RNA but not to AFF4 and enforces repression of HIV transcription
Retrovirology (2014)
-
The structure and substrate specificity of human Cdk12/Cyclin K
Nature Communications (2014)
-
Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition
Nature Communications (2012)