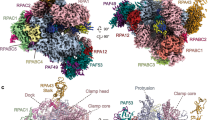
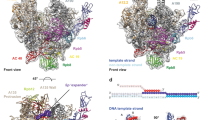
Abstract
To study how RNA polymerase II translocates after nucleotide incorporation, we prepared elongation complex crystals in which pre- and post-translocation states interconvert. Crystal soaking with the inhibitor α-amanitin locked the elongation complex in a new state, which was refined at 3.4-Å resolution and identified as a possible translocation intermediate. The DNA base entering the active site occupies a 'pretemplating' position above the central bridge helix, which is shifted and occludes the templating position. A leucine residue in the trigger loop forms a wedge at the shifted bridge helix, but moves by 13 Å to close the active site during nucleotide incorporation. Our results support a Brownian ratchet mechanism that involves swinging of the trigger loop between open, wedged and closed positions, and suggest that α-amanitin impairs nucleotide incorporation and translocation by trapping the trigger loop and bridge helix.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gnatt, A.L., Cramer, P., Fu, J., Bushnell, D.A. & Kornberg, R.D. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292, 1876–1882 (2001).
Kettenberger, H., Armache, K.-J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16, 955–965 (2004).
Westover, K.D., Bushnell, D.A. & Kornberg, R.D. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 119, 481–489 (2004).
Wang, D., Bushnell, D.A., Westover, K.D., Kaplan, C.D. & Kornberg, R.D. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954 (2006).
Cramer, P., Bushnell, D.A. & Kornberg, R.D. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science 292, 1863–1876 (2001).
Vassylyev, D.G. et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417, 712–719 (2002).
Bar-Nahum, G. et al. A ratchet mechanism of transcription elongation and its control. Cell 120, 183–193 (2005).
Epshtein, V. et al. Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol. Cell 10, 623–634 (2002).
Tuske, S. et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell 122, 541–552 (2005).
Artsimovitch, I. et al. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell 122, 351–363 (2005).
Bushnell, D.A., Cramer, P. & Kornberg, R.D. Structural basis of transcription: α-amanitin-RNA polymerase II cocrystal at 2.8 Å resolution. Proc. Natl. Acad. Sci. USA 99, 1218–1222 (2002).
Gong, X.Q., Nedialkov, Y.A. & Burton, Z.F. α-amanitin blocks translocation by human RNA polymerase II. J. Biol. Chem. 279, 27422–27427 (2004).
Vassylyev, D.G. et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature 448, 163–168 (2007).
Cramer, P. Gene transcription: extending the message. Nature 448, 142–143 (2007).
Landick, R. Active-site dynamics in RNA polymerases. Cell 116, 351–353 (2004).
Kashkina, E. et al. Multisubunit RNA polymerases melt only a single DNA base pair downstream of the active site. J. Biol. Chem. 282, 21578–21582 (2007).
Vassylyev, D.G., Vassylyeva, M.N., Perederina, A., Tahirov, T.H. & Artsimovitch, I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162 (2007).
Naji, S., Bertero, M.G., Spitalny, P., Cramer, P. & Thomm, M. Structure function analysis of the RNA polymerase cleft loops elucidates initial transcription, DNA unwinding and RNA displacement. Nucleic Acids Res. 36, 676–687 (2007).
Campbell, E.A. et al. Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase. EMBO J. 24, 674–682 (2005).
Toulokhonov, I., Zhang, J., Palangat, M. & Landick, R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell 27, 406–419 (2007).
Sousa, R. Machinations of a Maxwellian demon. Cell 120, 155–156 (2005).
Damsma, G.E., Alt, A., Brueckner, F., Carell, T. & Cramer, P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat. Struct. Mol. Biol. 14, 1127–1133 (2007).
Temiakov, D. et al. Structural basis for substrate selection by T7 RNA polymerase. Cell 116, 381–391 (2004).
Yin, Y.W. & Steitz, T.A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116, 393–404 (2004).
Cramer, P. Common structural features of nucleic acid polymerases. Bioessays 24, 724–729 (2002).
Abbondanzieri, E.A., Greenleaf, W.J., Shaevitz, J.W., Landick, R. & Block, S.M. Direct observation of base-pair stepping by RNA polymerase. Nature 438, 460–465 (2005).
Galburt, E.A. et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature 446, 820–823 (2007).
Chafin, D.R., Guo, H. & Price, D.H. Actions of α-amanitin during pyrophosphoryolysis and elongation by RNA polymerase II. J. Biol. Chem. 270, 19114–19119 (1995).
Rudd, M.D. & Luse, D.S. Amanitin greatly reduces the rate of transcription by RNA polymerase II ternary complexes but fails to inhibit some transcript cleavage modes. J. Biol. Chem. 271, 21549–21558 (1996).
Wienland, T. & Faulstich, H. Fifty years of amanitin. Experientia 47, 1186–1193 (1991).
Zanotti, G., Petersen, G. & Wieland, T. Structure-toxicity relationships in the amatoxin series. Int. J. Pept. Protein Res. 40, 551–558 (1992).
Armache, K.-J., Kettenberger, H. & Cramer, P. Architecture of the initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA 100, 6964–6968 (2003).
Brueckner, F., Hennecke, U., Carell, T. & Cramer, P. CPD damage recognition by transcribing RNA polymerase II. Science 315, 859–862 (2007).
Broennimann, E.F. et al. The PILATUS 1M detector. J. Synchrotron Radiat. 13, 120–130 (2006).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C. & Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 (2005).
Brunger, A.T. Version 1.2 of the Crystallography and NMR system. Nat. Protocols 2, 2728–2733 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Gerber, P.R. & Muller, K. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J. Comput. Aided Mol. Des. 9, 251–268 (1995).
Armache, K.-J., Mitterweger, S., Meinhart, A. & Cramer, P. Structures of complete RNA polymerase II and its subcomplex Rpb4/7. J. Biol. Chem. 280, 7131–7134 (2005).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Acknowledgements
We thank D. Kostrewa and other members of the Cramer laboratory for help. We thank D. Temiakov for discussions. F.B. was supported by the Nanosystems Initiative Munich (NIM) and the Elitenetzwerk Bayern (ENB). P.C. was supported by the Deutsche Forschungsgemeinschaft, the Sonderforschungsbereich SFB646, the Transregio 5 Chromatin, the EU research grant network 3D Repertoire and the Fonds der Chemischen Industrie. Part of this work was performed at the Swiss Light Source (SLS) at the Paul Scherrer Institut, Villigen, Switzerland.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brueckner, F., Cramer, P. Structural basis of transcription inhibition by α-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol 15, 811–818 (2008). https://doi.org/10.1038/nsmb.1458
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1458
This article is cited by
-
Exploring fungal RiPPs from the perspective of chemical ecology
Fungal Biology and Biotechnology (2022)
-
The occurrence of ansamers in the synthesis of cyclic peptides
Nature Communications (2022)
-
Incorporation of CENP-A/CID into centromeres during early Drosophila embryogenesis does not require RNA polymerase II–mediated transcription
Chromosoma (2022)
-
Liver transcriptome analyses of acute poisoning and recovery of male ICR mice exposed to the mushroom toxin α-amanitin
Archives of Toxicology (2022)
-
Nanoscopic subcellular imaging enabled by ion beam tomography
Nature Communications (2021)