Abstract
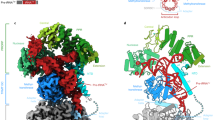
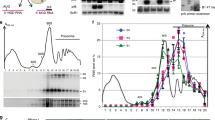
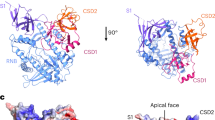
The maturation and stability of RNA transcripts is controlled by a combination of endo- and exoRNases. RNase J is unique, as it combines an RNase E–like endoribonucleolytic and a 5′-to-3′ exoribonucleolytic activity in a single polypeptide. The structural basis for this dual activity is unknown. Here we report the crystal structures of Thermus thermophilus RNase J and its complex with uridine 5′-monophosphate. A binding pocket coordinating the phosphate and base moieties of the nucleotide in the vicinity of the catalytic center provide a rationale for the 5′-monophosphate–dependent 5′-to-3′ exoribonucleolytic activity. We show that this dependence is strict; an initial 5′-PPP transcript cannot be degraded exonucleolytically from the 5′-end. Our results suggest that RNase J might switch promptly from endo- to exonucleolytic mode on the same RNA, a property that has important implications for RNA metabolism in numerous prokaryotic organisms and plant organelles containing RNase J orthologs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dreyfus, M. & Joyce, S. The interplay between translation and mRNA decay in procaryotes. in Translational Mechanisms (eds. Lapointe, J. & Brakier-Gingras, L.) 165–183 (Landes Bioscience, Georgetown, Texas, 2002).
Condon, C. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67, 157–174 (2003).
Condon, C. & Putzer, H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 30, 5339–5346 (2002).
Agaisse, H. & Lereclus, D. STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 20, 633–643 (1996).
Bechhofer, D.H. & Dubnau, D. Induced mRNA stability in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 84, 498–502 (1987).
Hambraeus, G., Karhumaa, K. & Rutberg, B. A 5′ stem-loop and ribosome binding site but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology 148, 1795–1803 (2002).
Glatz, E., Nilsson, R.-P., Rutberg, L. & Rutberg, B. A dual role for the Bacillus subtilis leader and the GlpP protein in the regulated expression of glpD: antitermination and control of mRNA stability. Mol. Microbiol. 19, 319–328 (1996).
Even, S. et al. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 33, 2141–2152 (2005).
Mathy, N. et al. 5′-to-3′ Exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129, 681–692 (2007).
Condon, C., Putzer, H., Luo, D. & Grunberg-Manago, M. Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J. Mol. Biol. 268, 235–242 (1997).
Bouvet, P. & Belasco, J.G. Control of RNase E–mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature 360, 488–491 (1992).
Tock, M.R., Walsh, A.P., Carroll, G. & McDowall, K.J. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′- end–dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 275, 8726–8732 (2000).
Mackie, G.A. Ribonuclease E is a 5′-end–dependent endonuclease. Nature 395, 720–723 (1998).
Jiang, X. & Belasco, J.G. Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc. Natl. Acad. Sci. USA 101, 9211–9216 (2004).
Callebaut, I., Moshous, D., Mornon, J.P. & de Villartay, J.P. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 30, 3592–3601 (2002).
Mandel, C.R. et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444, 953–956 (2006).
Ishikawa, H., Nakagawa, N., Kuramitsu, S. & Masui, R. Crystal structure of TTHA0252 from Thermus thermophilus HB8, a RNA degradation protein of the metallo-beta-lactamase superfamily. J. Biochem. 140, 535–542 (2006).
Dominski, Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 42, 67–93 (2007).
Li de la Sierra-Gallay, I., Mathy, N., Pellegrini, O. & Condon, C. Structure of the ubiquitous 3′ processing enzyme RNase Z bound to transfer RNA. Nat. Struct. Mol. Biol. 13, 376–377 (2006).
de la Sierra-Gallay, I.L., Pellegrini, O. & Condon, C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature 433, 657–661 (2005).
Weaver, L.H. et al. Comparison of goose-type, chicken-type, and phage-type lysozymes illustrates the changes that occur in both amino acid sequence and three-dimensional structure during evolution. J. Mol. Evol. 21, 97–111 (1984).
Cormack, R.S. & Mackie, G.A. Structural requirements for the processing of Escherichia coli 5 S ribosomal RNA by RNase E in vitro. J. Mol. Biol. 228, 1078–1090 (1992).
Mackie, G.A. Secondary structure of the mRNA for ribosomal protein S20. Implications for cleavage by ribonuclease E. J. Biol. Chem. 267, 1054–1061 (1992).
Mackie, G.A. & Genereaux, J.L. The role of RNA structure in determining RNase E–dependent cleavage sites in the mRNA for ribosomal protein S20 in vitro. J. Mol. Biol. 234, 998–1012 (1993).
McDowall, K.J., Kaberdin, V.R., Wu, S.W., Cohen, S.N. & Lin-Chao, S. Site-specific RNase E cleavage of oligonucleotides and inhibition by stem-loops. Nature 374, 287–290 (1995).
Callaghan, A.J. et al. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437, 1187–1191 (2005).
Choonee, N., Even, S., Zig, L. & Putzer, H. Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 35, 1578–1588 (2007).
Ryan, K., Calvo, O. & Manley, J.L. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10, 565–573 (2004).
Moshous, D. et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105, 177–186 (2001).
Ma, Y., Pannicke, U., Schwarz, K. & Lieber, M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108, 781–794 (2002).
Dominski, Z., Yang, X.C., Purdy, M., Wagner, E.J. & Marzluff, W.F.A. CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 25, 1489–1500 (2005).
Ma, Y. et al. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J. Biol. Chem. 280, 33839–33846 (2005).
Soubeyrand, S. et al. Artemis phosphorylated by DNA-dependent protein kinase associates preferentially with discrete regions of chromatin. J. Mol. Biol. 358, 1200–1211 (2006).
Niewolik, D. et al. DNA-PKcs dependence of Artemis endonucleolytic activity, differences between hairpins and 5′ or 3′ overhangs. J. Biol. Chem. 281, 33900–33909 (2006).
Taghbalout, A. & Rothfield, L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc. Natl. Acad. Sci. USA 104, 1667–1672 (2007).
Vanzo, N.F. et al. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12, 2770–2781 (1998).
Celesnik, H., Deana, A. & Belasco, J.G. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell 27, 79–90 (2007).
Muhlrad, D., Decker, C.J. & Parker, R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ → 3′ digestion of the transcript. Genes Dev. 8, 855–866 (1994).
Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 (1968).
Collaborative Computational Project, Number 4. The CCP 4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Kabsch, W.J. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Uson, I. & Sheldrick, G.M. Advances in direct methods for protein crystallography. Curr. Opin. Struct. Biol. 9, 643–648 (1999).
Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Perrakis, A., Morris, R. & Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 (1999).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Vagin, A.A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 (2004).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
DeLano, W.L. The PyMOL Molecular Graphics System. (DeLano Scientific, Palo Alto, California, 2002).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004).
Acknowledgements
We thank J. Plumbridge, M. Dreyfus, C. Condon and D. Picot for discussions, and Véronique Norman for help with the crystallization assays. We acknowledge the European Synchrotron Radiation Facility for providing the synchrotron radiation facilities and would like to thank X. Thibault for assistance in using beamline ID14-4, D. Picot and L. Barucq for the collection of the RNase J–UMP data at beamline ID29, J.L. Popot for the use of crystallography facilities and X-ray generator at the Institut de Biologie Physico-Chimique, and V. Urlacher and J. Berenguer for their gift of T. thermophilus chromosomal DNA. This work was supported by the Université Paris 7-Denis Diderot, the CNRS (UPR9073), the Agence Nationale de la Recherche and the Conseil Régionale de l'Ile de France. A.J. was supported by a scholarship from the Iranian Ministry of Science.
Author information
Authors and Affiliations
Contributions
Protein samples for structural studies were prepared by L.Z. Functional experiments and cloning were carried out by A.J. and L.Z., and analyzed by H.P. and A.J. Crystallization, structure calculation and refinement were carried out by I.L.d.l.S.-G. The manuscript was written by H.P. and I.L.d.l.S.-G.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Table 1 (PDF 2887 kb)
Rights and permissions
About this article
Cite this article
de la Sierra-Gallay, I., Zig, L., Jamalli, A. et al. Structural insights into the dual activity of RNase J. Nat Struct Mol Biol 15, 206–212 (2008). https://doi.org/10.1038/nsmb.1376
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1376
This article is cited by
-
Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nature Communications (2023)
-
The Arabidopsis chloroplast RNase J displays both exo- and robust endonucleolytic activities
Plant Molecular Biology (2019)
-
Tracking the elusive 5′ exonuclease activity of Chlamydomonas reinhardtii RNase J
Plant Molecular Biology (2018)
-
The role of ribonucleases in regulating global mRNA levels in the model organism Thermus thermophilus HB8
BMC Genomics (2014)
-
Initiation of mRNA decay in bacteria
Cellular and Molecular Life Sciences (2014)