Abstract
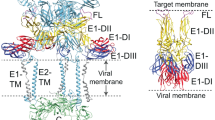
Improved technology for reconstructing cryo-electron microscopy (cryo-EM) images has now made it possible to determine secondary structural features of membrane proteins in enveloped viruses. The structure of mature dengue virus particles was determined to a resolution of 9.5 Å by cryo-EM and image reconstruction techniques, establishing the secondary structural disposition of the 180 envelope (E) and 180 membrane (M) proteins in the lipid envelope. The α-helical 'stem' regions of the E molecules, as well as part of the N-terminal section of the M proteins, are buried in the outer leaflet of the viral membrane. The 'anchor' regions of E and the M proteins each form antiparallel E-E and M-M transmembrane α-helices, leaving their C termini on the exterior of the viral membrane, consistent with the predicted topology of the unprocessed polyprotein. This is one of only a few determinations of the disposition of transmembrane proteins in situ and shows that the nucleocapsid core and envelope proteins do not have a direct interaction in the mature virus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Werten, P.J.L. et al. Progress in the analysis of membrane protein structure and function. FEBS Lett. 529, 65–72 (2002).
Unger, V.M., Kumar, N.M., Gilula, N.B. & Yeager, M. Projection structure of a gap junction membrane channel at 7 Å resolution. Nat. Struct. Biol. 4, 39–43 (1997).
Grigorieff, N., Ceska, T.A., Downing, K.H., Baldwin, J.M. & Henderson, R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 259, 393–421 (1996).
Cockburn, J.J.B., Bamford, J.K.H., Grimes, J.M., Bamford, D.H. & Stuart, D.I. Crystallization of the membrane-containing bacteriophage PRD1 in quartz capillaries by vapour diffusion. Acta Crystallogr. D 59, 538–540 (2003).
Lindenbach, B.D. & Rice, C.M. Flaviviridae: the viruses and their replication. In Fields Virology (eds. Knipe, D.M. & Howley, P.M.) 991–1041 (Lippincott Williams & Wilkins, Philadelphia, Pennsylvania, USA, 2001).
Burke, D.S. & Monath, T.P. Flaviviruses. In Fields Virology (eds. Knipe, D.M. & Howley, P.M.) 1043–1125 (Lippincott Williams & Wilkins, Philadelphia, Pennsylvania, USA, 2001).
Solomon, T. & Mallewa, M. Dengue and other emerging flaviviruses. J. Infect. 42, 104–115 (2001).
Hahn, Y.S. et al. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology 162, 167–180 (1988).
Elshuber, S., Allison, S.L., Heinz, F.X. & Mandl, C.W. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84, 183–191 (2003).
Kuhn, R.J. et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725 (2002).
Heinz, F.X. et al. The flavivirus envelope protein E: isolation of a soluble form from tick-borne encephalitis virus and its crystallization. J. Virol. 65, 5579–5583 (1991).
Rey, F.A., Heinz, F.X., Mandl, C., Kunz, C. & Harrison, S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375, 291–298 (1995).
Zhang, Y. et al. Structures of immature flavivirus particles. EMBO J. 22, 2604–2613 (2003).
Caspar, D.L.D. & Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27, 1–24 (1962).
Stiasny, K., Allison, S.L., Marchler-Bauer, A., Kunz, C. & Heinz, F.X. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 70, 8142–8147 (1996).
Allison, S.L., Schalich, J., Stiasny, K., Mandl, C.W. & Heinz, F.X. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75, 4268–4275 (2001).
Roehrig, J.T., Johnson, A.J., Hunt, A.R., Bolin, R.A. & Chu, M.C. Antibodies to dengue 2 virus E-glycoprotein syntheti(c peptides identify antigenic conformation. Virology 177, 668–675 (1990).
Bhardwaj, S., Holbrook, M., Shope, R.E., Barrett, A.D.T. & Watowich, S.J. Biophysical characterization and vector-specific antagonist activity of domain III of the tick-borne flavivirus envelope protein. J. Virol. 75, 4002–4007 (2001).
Crill, W.D. & Roehrig, J.T. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 75, 7769–7773 (2001).
Beasley, D.W.C. & Barrett, A.D.T. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J. Virol. 76, 13097–13100 (2002).
Allison, S.L., Stiasny, K., Stadler, K., Mandl, C.W. & Heinz, F.X. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73, 5605–5612 (1999).
Rossmann, M.G., Bernal, R. & Pletnev, S.V. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 136, 190–200 (2001).
Johnson, A.J., Guirakhoo, F. & Roehrig, J.T. The envelope glycoproteins of dengue 1 and dengue 2 viruses grown in mosquito cells differ in their utilization of potential glycosylation sites. Virology 203, 241–249 (1994).
Smith, G.W. & Wright, P.J. Synthesis of proteins and glycoproteins in dengue type 2 virus-infected vero and Aedes albopictus cells. J. Gen. Virol. 66, 559–571 (1985).
McGuffin, L.J., Bryson, K. & Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 (2000).
Zhang, W. et al. Placement of the structural proteins in Sindbis virus. J. Virol. 76, 11645–11658 (2002).
Jones, C.T. et al. Flavivirus capsid protein is a dimeric α-helical protein. J. Virol. 77, 7147–7179 (2003).
Mancini, E.J., Clarke, M., Gowen, B., Rutten, T. & Fuller, S.D. Cryo-electron microscopy reveals the functional anatomy of an enveloped virus, Semliki Forest virus. Mol. Cell 5, 255–266 (2000).
Lescar, J. et al. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105, 137–148 (2001).
Barth, B.U., Wahlberg, J.M. & Garoff, H. The oligomerization reaction of the Semliki Forest virus membrane protein subunits. J. Cell. Biol. 128, 283–291 (1995).
Lorenz, I.C., Allison, S.L., Heinz, F.X. & Helenius, A. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 76, 5480–5491 (2002).
Lee, S. et al. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 4, 531–541 (1996).
Skoging, U., Vihinen, M., Nilsson, L. & Liljeström, P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 4, 519–529 (1996).
Strauss, J.H., Strauss, E.G. & Kuhn, R.J. Budding of alphaviruses. Trends Microbiol. 3, 346–350 (1995).
Ng, M.L., Tan, S.H. & Chu, J.J. Transport and budding at two distinct sites of visible nucleocapsids of West Nile (Sarafend) virus. J. Med. Virol. 65, 758–764 (2001).
Strauss, J.H. & Strauss, E.G. Current advances in yellow fever research. In More About Microbe Hunters—Then and Now (eds. Koprowski, H. & Oldstone, M.B.A.) 113–137 (Medi-Ed Press, Bloomington, Illinois, USA, 1996).
Ji, Y., Marinescu, D.C., Zhang, W. & Baker, T.S. Orientation Refinement of Virus Structures with Unknown Symmetry (IEEE Computer Society, Los Alamitos, California, USA, 2003).
Rossmann, M.G. & Tao, Y. Cryo-electron microscopy reconstruction of partially symmetric objects. J. Struct. Biol. 125, 196–208 (1999).
Modis, Y, Ogata, S., Clements, D. & Harrison, S.C. A ligand–binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100, 6986–6991 (2003).
Acknowledgements
We thank E. Strauss for helpful discussions and R. Ashmore, C.R. Xiao, Y. Ji and D. Marinescu for the use of their various computer programs that were essential for calculating the image reconstruction. We are grateful for an equipment grant from the Keck Foundation. This work was supported by a US National Institutes of Health (NIH) Program Project grant to M.G.R., R.J.K. and T.S.B., by NIH grants to T.S.B. and J.H.S. and by a US National Science Foundation grant to T.S.B.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Zhang, W., Chipman, P., Corver, J. et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Mol Biol 10, 907–912 (2003). https://doi.org/10.1038/nsb990
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb990
This article is cited by
-
Development of a highly specific serodiagnostic ELISA for West Nile virus infection using subviral particles
Scientific Reports (2021)
-
Molecular characterization of structural protein genes of dengue virus serotype 1 epidemic in Yunnan, Southwest China, in 2018
Archives of Virology (2021)
-
Role of RNA-binding proteins during the late stages of Flavivirus replication cycle
Virology Journal (2020)
-
Comprehensive N-glycosylation mapping of envelope glycoprotein from tick-borne encephalitis virus grown in human and tick cells
Scientific Reports (2020)
-
Entry Inhibitors: Efficient Means to Block Viral Infection
The Journal of Membrane Biology (2020)