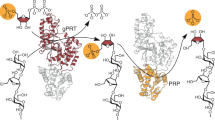
Abstract
Many bacteria and about 40,000 plant species form primary carbohydrate reserves based on fructan; these polymers of β-D-fructofuranose are thought to confer tolerance to drought and frost in plants. Microbial fructan, the β(2,6)-linked levan, is synthesized directly from sucrose by levansucrase, which is able to catalyze both sucrose hydrolysis and levan polymerization. The crystal structure of Bacillus subtilis levansucrase, determined to a resolution of 1.5 Å, shows a rare five-fold β-propeller topology with a deep, negatively charged central pocket. Arg360, a residue essential for polymerase activity, lies in a solvent-exposed site adjacent to the central pocket. Mutagenesis data and the sucrose-bound structure of inactive levansucrase E342A, at a resolution of 2.1 Å, strongly suggest that three conserved acidic side chains in the central pocket are critical for catalysis, and presumably function as nucleophile (Asp86) and general acid (Glu342), or stabilize the transition state (Asp247).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hendry, G. The ecological significance of fructan in a contemporary flora. New Phytol. 106, 201–216 (1987).
Cairns, A.J. Fructan biosynthesis in transgenic plants. J. Exp. Bot. 54, 549–567 (2003).
Vijn, I. & Smeekens, S. Fructan: more than a reserve carbohydrate? Plant Physiol. 120, 351–360 (1999).
Han, Y.W. Microbial levan. Adv. Appl. Microbiol. 35, 171–194 (1990).
Chambert, R., Treboul, G. & Dedonder, R. Kinetic studies of levansucrase of Bacillus subtilis. Eur. J. Biochem. 41, 285–300 (1974).
Coutinho, P.M. & Henrissat, B. in Recent Advances in Carbohydrate Bioengineering (eds. Gilbert, H.J., Davies, G., Henrissat, B. & Svensson, B.) 3–12 (The Royal Society of Chemistry, Cambridge, UK, 1999).
Rye, C.S. & Withers, S.G. Glycosidase mechanisms. Curr. Opin. Chem. Biol. 4, 573–580 (2000).
Chambert, R. & Gonzy-Treboul, G. Levansucrase of Bacillus subtilis: kinetic and thermodynamic aspects of transfructosylation processes. Eur. J. Biochem. 62, 55–64 (1976).
Hernandez, L. et al. Isolation and enzymic properties of levansucrase secreted by Acetobacter diazotrophicus SRT4, a bacterium associated with sugar cane. Biochem. J. 309, 113–118 (1995).
Song, D.D. & Jacques, N.A. Purification and enzymic properties of the fructosyltransferase of Streptococcus salivarius ATCC 25975. Biochem. J. 341, 285–291 (1999).
Chambert, R. & Gonzy-Treboul, G. Levansucrase of Bacillus subtilis. Characterization of a stabilized fructosyl–enzyme complex and identification of an aspartyl residue as the binding site of the fructosyl group. Eur. J. Biochem. 71, 493–508 (1976).
Bateman, A. et al. The Pfam protein families database. Nucleic Acids Res. 30, 276–280 (2002).
Song, D.D. & Jacques, N.A. Mutation of aspartic acid residues in the fructosyltransferase of Streptococcus salivarius ATCC 25975. Biochem. J. 344 (Part 1), 259–264 (1999).
Batista, F.R. et al. Substitution of Asp-309 by Asn in the Arg-Asp-Pro (RDP) motif of Acetobacter diazotrophicus levansucrase affects sucrose hydrolysis, but not enzyme specificity. Biochem. J. 337 (Part 3), 503–506 (1999).
Yanase, H. et al. Identification of functionally important amino acid residues in Zymomonas mobilis levansucrase. J. Biochem. 132, 565–572 (2002).
Chambert, R. & Petit-Glatron, M.F. Polymerase and hydrolase activities of Bacillus subtilis levansucrase can be separately modulated by site-directed mutagenesis. Biochem. J. 279, 35–41 (1991).
Kannan, R. et al. Molecular cloning and characterization of the extracellular sucrase gene (sacC) of Zymomonas mobilis. Arch. Microbiol. 163, 195–204 (1995).
Pons, T., Hernandez, L., Batista, F.R. & Chinea, G. Prediction of a common β-propeller catalytic domain for fructosyltransferases of different origin and substrate specificity. Protein Sci. 9, 2285–2291 (2000).
Naumoff, D.G. Conserved sequence motifs in levansucrases and bifunctional β-xylosidases and α-L-arabinases. FEBS Lett. 448, 177–179 (1999).
Nurizzo, D. et al. Cellvibrio japonicus α-L-arabinanase 43A has a novel five-blade β-propeller fold. Nat. Struct. Biol. 9, 665–668 (2002).
LeBrun, E. & van Rapenbusch, R. The structure of Bacillus subtilis levansucrase at 3.8 Å resolution. J. Biol. Chem. 255, 12034–12036 (1980).
Beisel, H.G., Kawabata, S., Iwanaga, S., Huber, R. & Bode, W. Tachylectin-2: crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus. EMBO J. 18, 2313–2322 (1999).
Paoli, M. Protein folds propelled by diversity. Prog. Biophys. Mol. Biol. 76, 103–130 (2001).
Petit-Glatron, M.F., Grajcar, L., Munz, A. & Chambert, R. The contribution of the cell wall to a transmembrane calcium gradient could play a key role in Bacillus subtilis protein secretion. Mol. Microbiol. 9, 1097–1106 (1993).
Harding, M.M. Geometry of metal–ligand interactions in proteins. Acta Crystallogr. D 57, 401–411 (2001).
Fülöp, V., Szeltner, Z. & Polgar, L. Catalysis of serine oligopeptidases is controlled by a gating filter mechanism. EMBO Rep. 1, 277–281 (2000).
Petit-Glatron, M.F., Benyahia, F. & Chambert, R. Secretion of Bacillus subtilis levansucrase: a possible two-step mechanism. Eur. J. Biochem. 163, 379–387 (1987).
Reddy, A. & Maley, F. Studies on identifying the catalytic role of Glu-204 in the active site of yeast invertase. J. Biol. Chem. 271, 13953–13957 (1996).
Dedonder, R. Levansucrase from Bacillus subtilis. Methods Enzymol. 86, 500–505 (1966).
Sierks, M.R. & Svensson, B. Energetic and mechanistic studies of glucoamylase using molecular recognition of maltose OH groups coupled with site-directed mutagenesis. Biochemistry 39, 8585–8592 (2000).
Vasella, A., Davies, G.J. & Böhm, M. Glycosidase mechanisms. Curr. Opin. Chem. Biol. 6, 619–629 (2002).
van den Elsen, J.M., Kuntz, D.A. & Rose, D.R. Structure of Golgi α-mannosidase II: a target for inhibition of growth and metastasis of cancer cells. EMBO J. 20, 3008–3017 (2001).
Chambert, R. & Petit-Glatron, M.F. Reversible thermal unfolding of Bacillus subtilis levansucrase is modulated by Fe3+ and Ca2+. FEBS Lett. 275, 61–64 (1990).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project Number 4. The CCP4 Suite of Programs for Protein Crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999).
Terwilliger, T.C. Automated structure solution, density modification and model building. Acta Crystallogr. D 58, 1937–1940 (2002).
Jones, T.A. & Thirup, S. Using known substructures in protein model building and crystallography. EMBO J. 5, 819–822 (1986).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. A 54, 905–921 (1998).
Winn, M.D., Isupov, M.N. & Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D 57, 122–133 (2001).
Hodel, A., Kim, S.-H. & Brünger, A. Model bias in crystal structures. Acta Crystallogr. A 48, 851–858 (1992).
Kleywegt, G.J. & Jones, T.A. Databases in protein crystallography. Acta Crystallogr. D 54, 1119–1131 (1998).
Carson, M. Ribbons. Methods Enzymol. 277, 493–505 (1997).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Engh, R.A. & Huber, R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 47, 392–400 (1991).
Acknowledgements
This work was supported by funds from the Royal Society and the School of Biosciences. G.M. is a recipient of the Adrian Brown Scholarship. We thank O.C. Mather for help with data collection and staff at ESRF for support at the beamlines. We used a computer cluster funded by the UK Medical Research Council and GlaxoWellcome, and received much valued technical advice from O.S. Smart, A.J. Pemberton and C. Cureton. We are indebted to R. Chambert for valuable discussions, to G. Waksman, J.B. Jackson, C.W. Wharton and S.A. White for comments on the manuscript and to L.M. Machesky and R.H. Insall for sharing their equipment.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Meng, G., Fütterer, K. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat Struct Mol Biol 10, 935–941 (2003). https://doi.org/10.1038/nsb974
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb974
This article is cited by
-
Preventive effect of Bacillus mojavensis levan against carbon tetrachloride and cisplatin toxicity: in vivo study
Environmental Science and Pollution Research (2021)
-
Exploring the sequence variability of polymerization-involved residues in the production of levan- and inulin-type fructooligosaccharides with a levansucrase
Scientific Reports (2019)
-
The K296-D320 region of recombinant levansucrase BA-SacB can affect the sensitivity of Escherichia coli host to sucrose
Annals of Microbiology (2019)
-
Understanding the transfer reaction network behind the non-processive synthesis of low molecular weight levan catalyzed by Bacillus subtilis levansucrase
Scientific Reports (2018)
-
Rational designed mutagenesis of levansucrase from Bacillus licheniformis 8-37-0-1 for product specificity study
Applied Microbiology and Biotechnology (2018)