Abstract
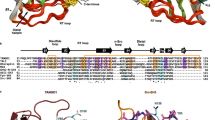
The Golgi-associated, γ-adaptin-related, ADP-ribosylation-factor binding proteins (GGAs) and adaptor protein (AP)-1 are adaptors involved in clathrin-mediated transport between the trans-Golgi network and endosomal system. The appendage domains of GGAs and the AP-1 γ-adaptin subunit are structurally homologous and have been proposed to bind to accessory proteins via interaction with short sequences containing phenylalanines and acidic residues. Here we present the structure of the human GGA1 appendage in complex with its cognate binding peptide from the p56 accessory protein (DDDDFGGFEAAETFD) as determined by X-ray crystallography. The interaction is governed predominantly by packing of the first two phenylalanine residues of the peptide with conserved basic and hydrophobic residues from GGA1. Additionally, several main chain hydrogen bonds cause the peptide to form an additional β-strand on the edge of the preexisting β-sheet of the protein. Isothermal titration calorimetry was used to assess the affinities of different peptides for the GGA and γ-appendage domains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kirchhausen, T. Adaptors for clathrin-mediated traffic. Annu. Rev. Cell Dev. Biol. 15, 705–732 (1999).
Boehm, M. & Bonifacino, J.S. Adaptins: the final recount. Mol. Biol. Cell 12, 2907–2920 (2001).
Scales, S.J., Gomez, M. & Kreis, T.E. Coat proteins regulating membrane traffic. Int. Rev. Cytol. 195, 67–144 (2000).
Collins, B.M., McCoy, A.J., Kent, H.M., Evans, P.R. & Owen, D.J. Molecular architecture and functional model of the endocytic AP-2 complex. Cell 109, 523–535 (2002).
Ricotta, D., Conner, S.D., Schmid, S.L., Figura, K.v. & Honing, S. Phosphorylation of the AP-2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J. Cell Biol. 156, 791–795 (2002).
Kent, H., McMahon, H., Evans, P., Benmerah, A. & Owen, D. γ-adaptin appendage domain. Structure and binding site for Eps15 and γ-synergin. Structure 10, 1139–1148 (2002).
Nogi, T. et al. Structural basis for the accessory protein recruitment by the γ-adaptin ear domain. Nat. Struct. Biol. 9, 527–531 (2002).
Boman, A.L., Zhang, C., Zhu, X. & Kahn, R.A. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell 11, 1241–1255 (2000).
Dell'Angelica, E.C. et al. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149, 81–94 (2000).
Hirst, J. et al. A family of proteins with γ-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell. Biol. 149, 67–80 (2000).
Poussu, A., Lohi, O. & Lehto, V.P. Vear, a novel Golgi-associated protein with VHS and γ-adaptin “ear” domains. J. Biol. Chem. 275, 7176–7183 (2000).
Takatsu, H., Yoshino, K. & Nakayama, K. Adaptor γ ear homology domain conserved in γ-adaptin and GGA proteins that interact with γ-synergin. Biochem. Biophys. Res. Commun. 271, 719–725 (2000).
Boman, A.L. GGA proteins: new players in the sorting game. J. Cell. Sci. 114, 3413–3418 (2001).
Black, M.W. & Pelham, H.R. Membrane traffic: how do GGAs fit in with the adaptors? Curr. Biol. 11, R460–462 (2001).
Robinson, M.S. & Bonifacino, J.S. Adaptor-related proteins. Curr. Opin. Cell Biol. 13, 444–453 (2001).
Doray, B., Ghosh P., Griffith, J., Geuze, H.J. & Kornfeld, S. Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science 297, 1700–1703 (2002).
Puertollano, R., Aguilar, R.C. & Gorshkova, I., Crouch, R.J. & Bonifacino, J.S. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292, 1712–1716 (2001).
Takatsu, H., Katoh, Y., Shiba, Y. & Nakayama, K. Golgi-localizing, γ-adaptin ear homology domain, ADP-ribosylation factor-binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J. Biol. Chem. 276, 28541–28545 (2001).
Zhu, Y., Doray, B., Poussu, A., Lehto, V.P. & Kornfeld, S. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science 292, 1716–1718 (2001).
Doray, B., Bruns, K., Ghosh, P. & Kornfeld, S. Interaction of the cation-dependent mannose 6-phosphate receptor with GGA proteins. J. Biol. Chem. 277, 18477–18482 (2002).
Misra, S., Puertollano, R., Kato, Y., Bonifacino, J.S. & Hurley, J.H. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415, 933–937 (2002).
Shiba, T. et al. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature 415, 937–941 (2002).
Puertollano, R., Randazzo, P.A., Presley, J.F., Hartnell, L.M. & Bonifacino, J.S. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell 105, 93–102 (2001).
Takatsu, H., Yoshino, K., Toda, K. & Nakayama, K. GGA proteins associate with Golgi membranes through interaction between their GGAH domains and ADP-ribosylation factors. Biochem. J. 365, 369–378 (2002).
Zhdankina, O., Strand, N.L., Redmond, J.M. & Boman, A.L. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast 18, 1–18 (2001).
Collins, B.M., Watson, P.J. & Owen, D.J. The structure of the GGA1 GAT domain reveals the molecular basis for ARF binding and membrane association of GGAs. Dev. Cell 4, 321–332 (2003).
Costaguta, G., Stefan, C.J., Bensen, E.S., Emr, S.D. & Payne, G.S. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell 12, 1885–1896 (2001).
Lui, W.W.Y. et al. Binding partners for the COOH-terminal appendage domains of the GGAs and γ-adaptin. Mol. Biol. Cell 14, 2385–2398 (2003).
Mattera, R., Arighi, C.N., Lodge, R., Zerial, M. & Bonifacino, J.S. Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J. 22, 78–88 (2003).
Page, L.J., Sowerby, P.J., Lui, W.W. & Robinson, M.S. γ-synergin: an EH domain-containing protein that interacts with γ-adaptin. J. Cell Biol. 146, 993–1004 (1999).
Kalthoff, C., Groos, S., Kohl, R., Mahrhold, S. & Ungewickell, E.J. Clint: a novel clathrin-binding ENTH-domain protein at the Golgi. Mol. Biol. Cell 13, 4060–4073 (2002).
Wasiak, S. et al. Entoprotin: a novel clathrin-associated protein identified through subcellular proteomics. J. Cell Biol. 158, 855–862 (2002).
Mills, I.G. et al. EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J. Cell Biol. 160, 213–222 (2003).
Hirst, J., Motley, A., Harasaki, K., Chew, S.Y.P. & Robinson, M.S. EpsinR: an ENTH domain containing protein that interacts with AP-1. Mol. Biol. Cell 14, 625–641 (2003).
Duncan, M.C., Costaguta, G. & Payne, G.S. Yeast epsin-related proteins required for Golgi-endosome traffic define a γ-adaptin ear-binding motif. Nat. Cell Biol. 5, 77–81 (2003).
Owen, D.J. & Evans, P.R. A structural explanation for the recognition of tyrosine based endocytic signals. Science 282, 1327–1332 (1998).
Brett, T.J., Traub, L.M. & Fremont, D.H. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure 10, 797–809 (2002).
Boll, W. et al. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 15, 5789–5795 (1996).
Harrison, S.C. Peptide-surface association: the case of PDZ and PTB domains. Cell 86, 341–343 (1996).
Munro, S. & Nichols, B.J. The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 9, 377–380 (1999).
Kjer-Nielsen, L., Teasdale, R.D., van Vliet, C. & Gleeson, P.A. A novel Golgi-localization domain shared by a class of coiled-coil peripheral membrane proteins. Curr. Biol. 9, 377–380 (1999).
Miller, G.J., Mattera, R., Bonifacino, J.S. & Hurley, J.H. Recognition of accessory protein motifs by the γ-adaptin ear domain of GGA3. Nat. Struct. Biol. advance online publication 13 July 2003 doi:10.1038/nsb953.
Aridor, M. & Traub, L.M. Cargo selection in vesicular transport: the making and breaking of a coat. Traffic 3, 537–546 (2002).
Powell, H.R., The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D 55, 1690–1695 (1999).
Collaborative Computational Project 4 (CCP4). The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Navaza, J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D 57, 1367–1372 (2001).
Murshudov, G.N., Vagin, A.A., Lebedev, A., Wilson, K.S. & Dodson, E.J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D 55, 247–255 (1999).
Jones, T.A., Zou, J.Y., Cowen, S.W. & Kjeldgaard, M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Diederichs, K. & Karplus, P.A. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275 (1997).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Acknowledgements
We thank H. Kent for the γ-appendage DNA, J. Hurley and associates for sharing information prior to publication and M. Noble for helpful discussions. G.J.K.P. is funded by a Marie Curie Fellowship of the European Union. D.J.O. is funded by a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science. M.S.R. is funded by a Wellcome Trust Principal Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Collins, B., Praefcke, G., Robinson, M. et al. Structural basis for binding of accessory proteins by the appendage domain of GGAs. Nat Struct Mol Biol 10, 607–613 (2003). https://doi.org/10.1038/nsb955
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb955
This article is cited by
-
The exomer cargo adaptor structure reveals a novel GTPase-binding domain
The EMBO Journal (2012)
-
The NECAP PHear domain increases clathrin accessory protein binding potential
The EMBO Journal (2007)
-
Life of a clathrin coat: insights from clathrin and AP structures
Nature Reviews Molecular Cell Biology (2006)
-
The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain
Nature Structural & Molecular Biology (2006)
-
γ-BAR, a novel AP-1-interacting protein involved in post-Golgi trafficking
The EMBO Journal (2005)