Abstract
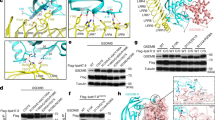
The cell death–inducing serine protease granzyme A (GzmA) has a unique disulfide-linked quaternary structure. The structure of human GzmA bound to a tripeptide CMK inhibitor, determined at a resolution of 2.4 Å, reveals that the oligomeric state contributes to substrate selection by limiting access to the active site for potential macromolecular substrates and inhibitors. Unlike other serine proteases, tetrapeptide substrate preferences do not correlate well with natural substrate cleavage sequences. This suggests that the context of the cleavage sequence within a macromolecular substrate imposes another level of selection not observed with the peptide substrates. Modeling of inhibitors bound to the GzmA active site shows that the dimer also contributes to substrate specificity in a unique manner by extending the active-site cleft. The crystal structure, along with substrate library profiling and mutagenesis, has allowed us to identify and rationally manipulate key components involved in GzmA substrate specificity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Russell, J.H. & Ley, T.J. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 20, 323–370 (2002).
Kam, C.M., Hudig, D. & Powers, J.C. Granzymes (lymphocyte serine proteases): characterization with natural and synthetic substrates and inhibitors. Biochem. Biophys. Acta 1477, 307–323 (2000).
Masson, D. & Tschopp, J. A family of serine esterase in lytic granules of cytolytic T lymphocytes. Cell 49, 679–685 (1987).
Shresta, S., Graubert, T.A., Thomas, D.A., Raptis, S.Z. & Ley, T.J. Granzyme A initiates an alternative pathway for granule-mediated apoptosis. Immunity 10, 595–605 (1999).
Beresford, P.J., Xia, Z., Greenberg, A.H. & Lieberman, J. Granzyme A loading induces rapid cytolysis and a novel form of DNA damage independently of caspase activation. Immunity 10, 585–594 (1999).
Beresford, P.J. et al. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J. Biol. Chem. 276, 43285–43293 (2001).
Fan, Z., Beresford, P.J., Oh, D.Y., Zhang, D. & Lieberman, J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nulceosome assembly protein SET is its inhibitor. Cell 112, 659–672 (2003).
Irmler, M. et al. Granzyme A is an interleukin 1 β-converting enzyme. J. Exp. Med. 181, 1917–1922 (1995).
Suidan, H.S. et al. Granzyme A released upon stimulation of cytotoxic T lymphocytes activates the thrombin receptor on neuronal cells and astroctyes. Proc. Natl. Acad. Sci. USA 91, 8112–8116 (1994).
Zhang, D., Beresford, P.J., Greenberg, A.H. & Lieberman, J. Granzymes A and B directly cleave lamins and disrupt the nuclear lamina during granule-mediated cytolysis. Proc. Natl. Acad. Sci. USA 98, 5746–5751 (2001).
Zhang, D. et al. Induction of rapid histone degradation by the cytotoxic T lymphocyte protease Granzyme A. J. Biol. Chem. 276, 3683–3690 (2001).
Fan, Z., Beresford, P.J., Zhang, D. & Lieberman, J. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol. Cell. Biol. 22, 2810–2820 (2002).
Fan, Z. et al. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat. Immunol. 4, 145–153 (2003).
Odake, S. et al. Human and murine cytotoxic T lymphocyte serine proteases subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry 30, 2217–2227 (1991).
Pasternack, M.S., Bleier, K.J. & McInerney, T.N. Granzyme A binding to target cell proteins. Granzyme A binds to and cleaves nucleolin in vitro. J. Biol. Chem. 266, 14703–14708 (1991).
Masson, D., Zamai, M. & Tschopp, J. Identification of granzyme A isolated from cytotoxic T-lymphocyte-granules as one of the proteases encoded by CTL-specific genes. FEBS Lett. 208, 84–88 (1986).
Schechter, I. & Berger, A. On the active site of protease. 3. Mapping the active of papain; specific peptide inhibitors of papain. Biochem. Biophys. Res. Comm. 32, 898–902 (1968).
Thornberry, N.A. et al. A combinatorial approach defines specificity of members of the caspase family and granzyme B. Functional relationships estabilshed for key mediators of apoptosis. J. Biol. Chem. 272, 17907–17911 (1997).
Harris, J.L. et al. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc. Natl. Acad. Sci. USA 97, 7754–7759 (2000).
Backes, B.J., Harris, J.L., Leonetti, F., Craik, C.S. & Ellman, J.A. Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat. Biotechnol. 18, 187–193 (2000).
Kim, S., Narayana, S.V. & Volanakis, J.V. Crystal structure of a complement factor D mutant expressing enhanced catalytic activity. J. Biol. Chem. 270, 24399–24405 (1995).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Galvin, J.P. et al. Apoptosis induced by granzyme B-glycosaminoglycan complexes: implications for granule-mediated apoptosis in vivo. J. Immunol. 164, 5345–5350 (1999).
Poe, M. et al. Human cytotoxic lymphocyte tryptase. Its purifcation from granules and the characterization of inhibitor and substrate specificity. J. Biol. Chem. 263, 13215–13222 (1988).
Barrios, A.M. & Craik, C.S. Scanning the prime-site substrate specificity of proteolytic enzymes: A novel assay based on ligand-enhanced lanthanide ion fluorescence. Bioorg. Med. Chem. Lett. 12, 3619–3623 (2002).
Simon, M.M., Hoschutzky, H., Fruth, U., Simon, H.G. & Kramer, M.D. Purification and characterization of a T cell specific serine proteinase (TSP-1) from cloned cytolytic T lymphocytes. EMBO J. 5, 3267–3274 (1986).
Fruth, U. et al. A novel serine proteinase (HuTSP) isolated from a cloned human CD8+ cytolytic T cell line is expressed and secreted by activated CD4+ and CD8+ lymphocytes. Eur. J. Immunol. 17, 1625–1633 (1987).
Krahenbuhl, O. et al. Characterization of granzymes A and B isolated from granules of cloned human cytotoxic T lymphocytes. J. Immunol. 141, 3471–3477 (1988).
Murphy, M.E. et al. Comparative molecular model building of two serine proteinases from cytotoxic T lymphocytes. Proteins 4, 190–204 (1988).
Harris, J.L., Peterson, E.P., Hudig, D., Thornberry, N.A. & Craik, C.S. Definition and redesign of the extended substrate specificity of granzyme B. J. Biol. Chem. 273, 27364–27373 (1998).
Waugh, S.M., Harris, J.L., Fletterick, R. & Craik, C.S. The structure of the pro-apoptotic protease granzyme B reveals the molecular determinants of its specificity. Nat. Struct. Biol. 7, 762–765 (2000).
Earnshaw, W.C., Martins, L.M. & Kaufmann, S.H. Mammalian caspases: Structure, activation, substratea and functions during apoptosis. Annu. Rev. Biochem. 68, 383–424 (1999).
Pereira, P.J. et al. Human β-tryptase is a ring-like tetramer with active sites facing a central pore. Nature 392, 306–311 (1998).
Perona, J.J., Tsu, C.A., Craik, C.S. & Fletterick, R.J. Crystal structure of an ecotin-collagenase complex suggest a model for recognition and cleavage of the collagen triple helix. Biochemistry 36, 5381–5392 (1997).
Eggers, C.T., Wang, S.X., Fletterick, R. & Craik, C.S. The role of ecotin dimerization in protease inhibition. J. Mol. Biol. 308, 975–991 (2001).
Wang, S.X., Esmon, C.T. & Fletterick, R.J. Crystal structure of thrombin-ecotin reveals conformational changes and extended interactions. Biochemistry 40, 10038–10046 (2001).
Tsuzuki, S. et al. Purification and identification of a binding protein for pancreatic secretory trypsin inhbitor: a novel role of the inhibitor as an anti-granzyme A. Biochem. J. 372, 227–233 (2003).
Hink-Schauer, C., Estebanez-Perpina, E., Bode, W. & Jenne, D.E. Crystal structure of the apoptosis-inducing human granzyme A dimer. Nat. Struct. Biol. 10, 535–540 (2003).
Jackson, D.S. et al. Synthesis and evaluation of diphenyl phosphonate esters as inhibitors of the trypsin-like granzymes A and K and mast cell tryptase. J. Med. Chem. 41, 2289–2301 (1998).
Gill, S.C. & von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 (1989).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Kissinger, C.R., Gehlhaar, D.K. & Fogel, D.B. Rapid Automated Molecular Replacement By Evolutionary Search. Acta Crystallogr. D 55, 484–491 (1999).
Cowtan, K. 'dm': An automated procedure for phase improvement by density modification. Joint CCP4/ESF-EACBM Newsletter on Prot. Crystallogr. 31, 34–38 (1994).
Lamzin, V.S. & Wilson, K.S. Automated refinement of protein models. Acta Crystallogr. D 49, 129–149 (1993).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Murshudov, G., Vagin, A. & Dodson, E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Peitsch, M.C. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24, 274–279 (1996).
Acknowledgements
The authors would like to express thanks to J. Holton for technical assistance during data collection at the ALS beam line 8.3.1. We would also like to acknowledge the Macromolecular Structure Group at the University of California, San Francisco for computational support. This work was supported in part by a US National Institutes of Health post-doctoral training grant to J.K.B. and grants from the US National Institutes of Health to C.S.C. and R.J.F.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Bell, J., Goetz, D., Mahrus, S. et al. The oligomeric structure of human granzyme A is a determinant of its extended substrate specificity. Nat Struct Mol Biol 10, 527–534 (2003). https://doi.org/10.1038/nsb944
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb944
This article is cited by
-
Identification and annotation of bovine granzyme genes reveals a novel granzyme encoded within the trypsin-like locus
Immunogenetics (2018)
-
Proteolytic clipping of histone tails: the emerging role of histone proteases in regulation of various biological processes
Molecular Biology Reports (2014)
-
Expression profile of novel members of the rat mast cell protease (rMCP)-2 and (rMCP)-8 families, and functional analyses of mouse mast cell protease (mMCP)-8
Immunogenetics (2007)
-
Granzyme A, which causes single‐stranded DNA damage, targets the double‐strand break repair protein Ku70
EMBO reports (2006)
-
Molecular characterization and expression of a granzyme of an ectothermic vertebrate with chymase-like activity expressed in the cytotoxic cells of Nile tilapia (Oreochromis niloticus)
Immunogenetics (2006)