Abstract
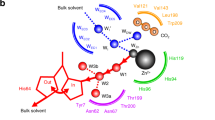
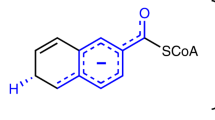
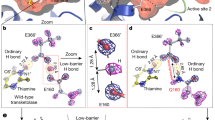
Zinc-dependent enzymes play important roles in many cellular processes. Assignment of their reaction mechanisms is often a subject of debate because the zinc ion is silent in several spectroscopic techniques. We have combined time-resolved X-ray absorption spectroscopy, pre-steady state kinetics and computational quantum chemistry to study the active site zinc ion of bacterial alcohol dehydrogenase during single substrate turnover. We detect a series of alternations in the coordination number and structure of the catalytic zinc ion with concomitant changes in metal–ligand bond distances. These structural changes are reflected in the effective charge of the metal ion. The present work emphasizes the flexibility of catalytic zinc sites during catalysis and provides novel mechanistic insights into alcohol dehydrogenase catalysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Auld, D.S. Zinc coordination sphere in biochemical zinc sites. Biometals 14, 271–313 (2001).
Karlin, K.D. Metalloenzymes, structural motifs, and inorganic models. Science 261, 701–708 (1993).
Oestreicher, E.G., Pereira, D.A. & Pinto, G.F. Steady-state kinetic mechanism of Thermoanaerobacter brockii alcohol dehydrogenase: a study discrimination between alternative kinetic models. J. Biotechnol. 46, 23–31 (1996).
al-Kassim, L.S. & Tsai, C.S. Studies of NADP+-preferred secondary alcohol dehydrogenase from Thermoanaerobium brockii. Biochem. Cell Biol. 68, 907–913 (1990).
Korkhin, Y. et al. NADP-dependent bacterial alcohol dehydrogenases: crystal structure, cofactor-binding and cofactor specificity of the ADHs of Clostridium beijerinckii and Thermoanaerobacter brockii. J. Mol. Biol. 278, 967–981 (1998).
Kleifeld, O. et al. Spectroscopic studies of inhibited alcohol dehydrogenase from Thermoanaerobacter brockii: proposed structure for the catalytic intermediate state. Biochemistry 39, 7702–7711 (2000).
Dworschack, R.T. & Plapp, B.V. Kinetics of native and activated isozymes of horse liver alcohol dehydrogenase. Biochemistry 16, 111–116 (1977).
Dunn, M.F., Biellmann, J.F. & Branlant, G. Roles of zinc ion and reduced coenzyme in horse liver alcohol dehydrogenase catalysis. The mechanism of aldehyde activation. Biochemistry 14, 3176–3182 (1975).
Makinen, M.W., Maret, W. & Yim, M.B. Neutral metal-bound water is the base catalyst in liver alcohol dehydrogenase. Proc. Natl. Acad. Sci. USA 80, 2584–2588 (1983).
Meijers, R. et al. On the enzymatic activation of NADH. J. Biol. Chem. 276, 9316–9321 (2001).
Dunn, M.F., MacGibbon, A.K.H. & Pease, K. Liver alcohol dehydrogenase:electrostatic strain-distortion effects facilitate hydride ion transfer. in Zinc Enzymes, Vol. 1 (eds. Bertini, I., Luchinat, C., Maret, W. & Zeppezauer, M.) 378–534 (Birkhauser Boston, Boston; 1986).
Eklund, H., Plapp, B.V., Samama, J.P. & Branden, C.I. Binding of substrate in a ternary complex of horse liver alcohol dehydrogenase. J. Biol. Chem. 257, 14349–14358 (1982).
Powers, L., Sessler, L., Woolery, G.L. & Chance, B. CO bond angle changes in photolysis of carboxymyoglobin. Biochemistry 23, 5519–5523 (1984).
Scheuring, E.M. et al. Time-resolved X-ray absorption spectroscopy of photoreduced base-off Cob(II)alamin compared to the Co(II) species in Clostridium thermoaceticum. J. Phys. Chem. 100, 3344–3348 (1996).
Chance, M.R. et al. Global mapping of structural solutions provided by the extended X-ray absorption fine structure ab initio code FEFF 6.01: structure of the cryogenic photoproduct of the myoglobin-carbon monoxide complex. Biochemistry 35, 9014–9023 (1996).
Riggs-Gelasco, P. et al. EXAFS characterization of the intermediate X generated during the assembly of the E. coli ribonucleotide reductase R2 diferric tyrosyl radical cofactor. J. Am. Chem. Soc. 120, 849–860 (1998).
Hwang, J. et al. A short Fe-Fe distance in peroxodiferric ferritin: control of Fe substrate versus cofactor decay? Science 287, 122–125 (2000).
Lee, S.K. et al. Nature of the intermediate formed in the reduction of O2 to H2O at the trinuclear copper cluster active site in native laccase. J. Am. Chem. Soc. 124, 6180–6193 (2002).
Fersht, A. The basic equations of enzyme kinetics. in Structure and Mechanism in Protein Science (eds. Julet, M.R. & Hadler, G.L.) 103–131 (W.H. Freeman, New York; 1999).
Frenkel, A., Kleifeld, O., Wasserman, S.R. & Sagi, I. Phase speciation by extended X-ray-absorption fine structure spectroscopy. J. Chem. Phys. 116, 9449–9456 (2002).
Zabinsky, S.I., Rehr, J.J., Ankudinov A., Albers R.C. & Eller M.J. Multiple scattering calculations of X-ray absorption spectra. Phys. Rev. B 52, 2995–3009 (1995).
Frieden, C. Slow transitions and hysteretic behavior in enzymes. Ann. Rev. Biochem. 48, 471–489 (1979).
Bianconi, A. XANES spectroscopy. in X-Ray Absorption, Vol. 92 (eds. Koningsberger, D.C. & Prins, R.) 573–662 (John Wiley & Sons, New York; 1988).
Maret, W. & Makinen, M.W. The pH variation of steady-state kinetic parameters of site-specific Co2+-reconstituted liver alcohol dehydrogenase a mechanistic probe for the assignment of metal-linked inoziations. J. Biol. Chem. 266, 20636–20644 (1991).
Cioslowski, J. A new population analysis based on atomic polar tensors. J. Am. Chem. Soc. 111, 8333–8336 (1989).
Reed, A.E., Curtiss, L.A. & Weinhold, F. Intermolecular interactions from a natural bond orbital, donor-acceptor viewpoint. Chem. Rev. 88, 899–926 (1988).
Ryde, U. On the role of Glu68 in alcohol dehydrogenase. Protein Sci. 4, 1124–1132 (1995).
Yang, Z.N., Bosron, W.F. & Hurley, T.D. Structure of human chi chi alcohol dehydrogenase: a glutathione-dependent formaldehyde dehydrogenase. J. Mol. Biol. 265, 330–343 (1997).
Bogin, O., Peretz, M. & Burstein, Y. Thermoanaerobacter brockii alcohol dehydrogenase: characterization of the active site metal and its ligand amino acids. Protein Sci. 6, 450–458 (1997).
Appleyard, R.J., Shuttleworth, W.A. & Evans, J.N. Time-resolved solid-state NMR spectroscopy of 5-enolpyruvylshikimate-3-phosphate synthase. Biochemistry 33, 6812–6821 (1994).
Stern, E.A., Newville, M., Ravel, B., Yacoby, Y. & Haskel, D. The UWXAFS analysis package: philosophy and details. Physica B 208-209, 117–120 (1995).
Rehr, J.J., Mustre de Leon, J., Zabinsky, S.I. & Albers, R.C. Theoretical X-ray absorption fine structure standards. J. Am. Chem. Soc. 113, 5135–5140 (1991).
Frisch, M.J. et al. Gaussian 98. (Rev. A.11) (Gaussian, Inc., Pittsburgh; 2001).
Acknowledgements
We thank M. Sullivan from the NSLS beam line X9B, K. Zhang and S. Stepanov from the APS the BioCat beamline and G. Kafri and E. Yavin from the Weizmann Institute for their help and technical advice. The authors are grateful to M.W. Makinen for critically reading this manuscript. This work was supported by the Israeli Science Foundation (I.S.) and by the Minerva Foundation (J.M.L.M.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kleifeld, O., Frenkel, A., Martin, J. et al. Active site electronic structure and dynamics during metalloenzyme catalysis. Nat Struct Mol Biol 10, 98–103 (2003). https://doi.org/10.1038/nsb889
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb889
This article is cited by
-
Rapid Freeze-Quench EPR Spectroscopy: Improved Collection of Frozen Particles
Applied Magnetic Resonance (2016)
-
Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins
Nature Communications (2014)
-
The improvement of stability, activity, and substrate promiscuity of glycerol dehydrogenase substituted by divalent metal ions
Biotechnology and Bioprocess Engineering (2013)
-
Correlated structural kinetics and retarded solvent dynamics at the metalloprotease active site
Nature Structural & Molecular Biology (2011)