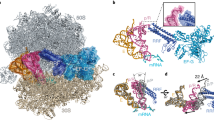
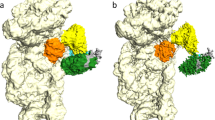
Abstract
The L1 protuberance of the 50S ribosomal subunit is implicated in the release/disposal of deacylated tRNA from the E site. The apparent mobility of this ribosomal region has thus far prevented an accurate determination of its three-dimensional structure within either the 50S subunit or the 70S ribosome. Here we report the crystal structure at 2.65 Å resolution of ribosomal protein L1 from Sulfolobus acidocaldarius in complex with a specific 55-nucleotide fragment of 23S rRNA from Thermus thermophilus. This structure fills a major gap in current models of the 50S ribosomal subunit. The conformations of L1 and of the rRNA fragment differ dramatically from those within the crystallographic model of the T. thermophilus 70S ribosome. Incorporation of the L1–rRNA complex into the structural models of the T. thermophilus 70S ribosome and the Deinococcus radiodurans 50S subunit gives a reliable representation of most of the L1 protuberance within the ribosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ban, N., Nissen, P., Hansen, J., Moore, P.B. & Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000).
Harms, J. et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107, 679–688 (2001).
Yusupov, M.M. et al. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 (2001).
Zimmermann, R.A. Interactions among protein and RNA components of the ribosome. in Ribosomes. Structure, Function and Genetics (eds. Chambliss, G. et al.) 135–169 (University Park Press, Baltimore; 1980).
Malhotra, A. et al. Escherichia coli 70 S ribosome at 15 Å resolution by cryo-electron microscopy: localization of fMet-tRNAfMet and fitting of L1 protein. J. Mol. Biol. 280, 103–116 (1998).
Agrawal, R.K. et al. Visualization of tRNA movements on the E. coli 70S ribosome during the elongation cycle. J. Cell Biol. 150, 447–460 (2000).
Moazed, D. & Noller, H.F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 57, 585–597 (1989).
Wower, J. et al. Transit of tRNA through the Escherichia coli ribosome. Cross-linking of the 3′ end of tRNA to specific nucleotides of the 23 S ribosomal RNA at the A, P, and E sites. J. Biol. Chem. 275, 37887–37894 (2000).
Kirillov, S.V., Wower, J., Hixson, S.H. & Zimmermann, R.A. Transit of tRNA through the Escherichia coli ribosome: cross-linking of the 3′ end of tRNA to ribosomal proteins at the P and E sites. FEBS Lett. 514, 60–66 (2002).
Gabashvili, I.S. et al. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100, 537–549 (2000).
Gomez-Lorenzo, M.G. et al. Tree-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J. 19, 2710–2718 (2000).
Baier, G. et al. Structural and functional equivalence between ribosomal proteins of Escherichia coli L1 and Methanococcus vannielii L6. Syst. Appl. Microbiol. 12, 119–126 (1989).
Hanner, M. et al. Autogenous translational regulation of the ribosomal MvaL1 operon in the archaebacterium Methanococcus vannielii. J. Bacteriol. 176, 409–418 (1994).
Nevskaya, N. et al. Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family. Structure 8, 363–371 (2000).
Nevskaya, N. et al. Structure of ribosomal protein L1 from Methanococcus thermolithotrophicus. Functionally important structural invariants on the L1 surface. Acta Crystallogr. D 58, 1023–1029 (2002).
Egebjerg, J., Christiansen, J. & Garret, R.A. Attachment sites of primary binding proteins L1, L2 and L23 on 23S ribosomal RNA of Escherichia coli. J. Mol. Biol. 222, 251–264 (1991).
Doring, T., Greuer, B. & Brimacombe, R. The three-dimensional folding of ribosomal RNA; localization of a series of intra-RNA cross-links in 23S RNA induced by treatment of Escherichia coli 50S ribosomal subunits with bis-(2-chloroethyl)-methylamine. Nucleic Acids Res. 19, 3517–3524 (1991).
Nikonov, S. et al. Crystal structure of the RNA binding ribosomal protein L1 from Thermus thermophilus. EMBO J. 15, 1350–1359 (1996).
Stanley, J., Sloof, P. & Ebel, J.-P. The binding site of ribosomal protein L1 from Escherichia coli on the 23S ribosomal RNA from Bacillus stearothermophilus. A possible base-pairing scheme differing from that proposed for Escherichia coli. Eur. J. Biochem. 85, 309–316 (1978).
Zimmermann, R.A., Thurlow, D.L., Finn, R.S., Marsh, T.L. & Ferrett, L.K. Conservation of specific protein-RNA interactions in ribosome evolution. in Genetics and Evolution of RNA Polymerase, tRNA and Ribosomes (eds. Osava, S., Ozeki, H., Uchida, H. & Yura, T.) 569–584 (University of Tokyo Press, Tokyo; 1980)
Gourse, R.L., Thurlow, D.L., Gerbi, S.A. & Zimmermann, R.A. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proc. Natl. Acad. Sci. USA 78, 2722–2726 (1981).
Baier, G., Piendl, W., Redl, B. & Stoffler, G. Structure, organization and evolution of the L1 equivalent ribosomal protein gene of the archaebacterium Methanococcus vannielii. Nucleic Acids Res. 18, 719–724 (1990).
Yusupov, M.M. & Spirin, A.S. Are there proteins between the ribosomal subunits? Hot tritium bombardment experiments. FEBS Lett. 197, 229–233 (1986).
Drygin, D. & Zimmermann, R.A. Magnesium ions mediate contacts between phosphoryl oxygens at positions 2122 and 2176 of the 23S rRNA and ribosomal protein L1. RNA 6, 1714–1726 (2000).
Kraft, A., Lutz, C., Lingenhel, A., Gröbner, P. & Piendl, W. Control of ribosomal protein L1 synthesis in mesophilic and thermophilic archaea. Genetics 152, 1363–1372 (1999).
Brinkmann, U., Mattes, R.E. & Buckel, P. High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene 85, 109–114 (1989).
Tishchenko, S. et al. Crystals of ribosomal protein L1 from a hyperthermophilic archaeon Methanococcus jannaschii. Biochem. Mol. Biol. Int. 45, 349–354 (1998).
Kabsch, W. Integration, scaling, space-group assignment and post refinement. XDS. in International Tables for Crystallography (eds. Rossmann, M.G. & Arnold, E.) F (Kluwer Academic Publishers, Dordrecht; 2001).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Zhou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Acknowledgements
This work was supported by the Russian Academy of Sciences and the Russian Foundation for Basic Research. The research of M.G. was supported in part by the International Research Scholar's award from the Howard Hughes Medical Institute. The research of W.P. was supported by the Austrian Science Fund and the Austrian-Russian Collaboration Program. The research of R.A.Z. was supported by a grant from the U.S. National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Nikulin, A., Eliseikina, I., Tishchenko, S. et al. Structure of the L1 protuberance in the ribosome. Nat Struct Mol Biol 10, 104–108 (2003). https://doi.org/10.1038/nsb886
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb886
This article is cited by
-
Studying the Properties of Domain I of the Ribosomal Protein L1: Incorporation into Ribosome and Regulation of the L1 Operon Expression
The Protein Journal (2015)
-
Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA
Nature (2013)
-
A hierarchical model for evolution of 23S ribosomal RNA
Nature (2009)
-
What recent ribosome structures have revealed about the mechanism of translation
Nature (2009)
-
The Key Role of Large Clusters of Polar Residues of RNA-Binding Proteins in the Formation of Complexes with RNA
Molecular Biology (2005)