Abstract
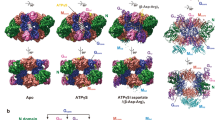
The structure of the cooperative hexameric enzyme ATP sulfurylase from Penicillium chrysogenum bound to its allosteric inhibitor, 3′-phosphoadenosine-5′-phosphosulfate (PAPS), was determined to 2.6 Å resolution. This structure represents the low substrate-affinity T-state conformation of the enzyme. Comparison with the high substrate-affinity R-state structure reveals that a large rotational rearrangement of domains occurs as a result of the R-to-T transition. The rearrangement is accompanied by the 17 Å movement of a 10-residue loop out of the active site region, resulting in an open, product release-like structure of the catalytic domain. Binding of PAPS is proposed to induce the allosteric transition by destabilizing an R-state-specific salt linkage between Asp 111 in an N-terminal domain of one subunit and Arg 515 in the allosteric domain of a trans-triad subunit. Disrupting this salt linkage by site-directed mutagenesis induces cooperative inhibition behavior in the absence of an allosteric effector, confirming the role of these two residues.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
MacRae, I. & Segel, I.H. Arch. Biochem. Biophys. 337, 17–26 (1997).
Renosto, F., Martin, R.L., Wailes, L.M., Daley, L.A. & Segel, I.H. J. Biol. Chem. 265, 10300–10308 (1990).
MacRae, I.J., Segel, I.H. & Fisher, A.J. Biochemistry 40, 6795–6804 (2001).
Foster, B.A. et al. J. Biol. Chem. 269, 19777–19786 (1994).
Monod, J., Wyman, J. & Changeux, J.-P. J. Mol. Biol. 12, 88–118 (1965).
Lipscomb, W.N. Adv. Enzymol. Relat. Areas Mol. Biol. 68, 67–151 (1994).
Deyrup, A.T., Singh, B., Krishnan, S., Lyle, S. & Schwartz, N.B. J. Biol. Chem. 274, 28929–28936 (1999).
Venkatachalam, K.V., Fuda, H., Koonin, E.V. & Strott, C.A. J. Biol. Chem. 274, 2601–2604 (1999).
Serrano, L. & Fersht, A.R. Nature 342, 296–299 (1989).
Beynon, J.D. et al. Biochemistry 40, 14509–14517 (2001).
Ullrich, T.C. & Huber, R. J. Mol. Biol. 313, 1117–1125 (2001).
Ullrich, T.C., Blaesse, M. & Huber, R. EMBO J. 20, 316–329 (2001).
MacRae, I.J., Hanna, E., Ho, J.D., Fisher, A.J. & Segel, I.H. J. Biol. Chem. 275, 36303–36310 (2000).
Medina, D.C., Hanna, E., MacRae, I.J., Fisher, A.J. & Segel, I.H. Arch. Biochem. Biophys. 393, 51–60 (2001).
Lu, G., Williams, M.K., Giroux, E.L. & Kantrowitz, E.R. Biochemistry 34, 13272–13277 (1995).
Dembowski, N.J., Newton, C.J. & Kantrowitz, E.R. Biochemistry 29, 3716–3723 (1990).
Williams, M.K., Stec, B. & Kantrowitz, E.R. J. Mol. Biol. 281, 121–134 (1998).
Williams, M.K. & Kantrowitz, E.R. Biochim. Biophys. Acta 1429, 249–258 (1998).
Sakash, J.B. & Kantrowitz, E.R. J. Biol. Chem. 275, 28701–28707 (2000).
Schmidheini, T., Mosch, H.U., Evans, J.N. & Braus, G. Biochemistry 29, 3660–3668 (1990).
Stevens, R.C. & Lipscomb, W.L. in Oxford Science Publications; Molecular Structures in Biology (eds Diamond, R., Koetzle, T.F., Prout, K. & Richardson, J.S.) 223–259 (Oxford University Press, Oxford; 1993).
Kantrowitz, E.R. & Lipscomb, W.N. Trends Biochem. Sci. 15, 53–59 (1990).
Horjales, E., Altamirano, M.M., Calcagno, M.L., Garratt, R.C. & Oliva, G. Structure 7, 527–537 (1999).
Brünger, A.T. et al. Acta Crystallogr. D 54, 905–921 (1998).
Kissinger, C.R., Gehlhaar, D.K. & Fogel, D.B. Acta Crystallogr. D 55, 484–491 (1999).
Brünger, A.T. Acta Crystallogr. D 49, 24–36 (1993).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Acta Crystallogr. A 47, 110–119 (1991).
Segel, I.H. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. (Wiley-Interscience, New York; 1993).
Acknowledgements
The research described in this report was supported by an NSF Research Grant to I.H.S. and A.J.F. and by facilities of the W.M. Keck Foundation Center for Structural Biology at the University of California, Davis. Some of the work reported here was performed at SSRL, which is operated by the Department of Energy, Office of Basic Energy Sciences. The SSRL Biotechnology Program is supported by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and by the Department of Energy, Office of Biological and Environmental Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
MacRae, I., Segel, I. & Fisher, A. Allosteric inhibition via R-state destabilization in ATP sulfurylase from Penicillium chrysogenum. Nat Struct Mol Biol 9, 945–949 (2002). https://doi.org/10.1038/nsb868
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb868