Abstract
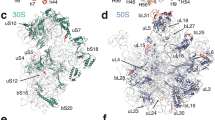
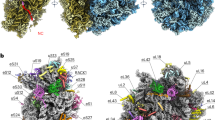
Understanding the structural basis of ribosomal function requires close comparison between biochemical and structural data. Although a large amount of biochemical data are available for the Escherichia coli ribosome, the structure has not been solved to atomic resolution. Using a new RNA homology procedure, we have modeled the all-atom structure of the E. coli 30S ribosomal subunit. We find that the tertiary structure of the ribosome core, including the A-, P- and E-sites, is highly conserved. The hypervariable regions in our structure, which differ from the structure of the 30S ribosomal subunit from Thermus thermophilus, are consistent with the cryo-EM map of the E. coli ribosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wimberly, B.T. et al. Nature 407, 327–339 (2000).
Schluenzen, F. et al. Cell 102, 615–623 (2000).
Green, R. & Noller, H.F. Annu. Rev Biochem 66, 679–716 (1997).
O'Connor, M. in The Ribosome: Structure, Function, Antibiotics, & Cellular Interactions (eds Garrett, R.A. et al.) 217–227 (ASM Press, Washington, D.C.; 2000).
Sanchez, R. & Sali, A. Curr. Opin. Struct. Biol. 7, 206–214 (1997).
Westhof, E. Theochem 105, 203–210 (1993).
Westhof, E. & Altman, S. Proc. Natl. Acad. Sci. USA 9, 5133–5137 (1994).
Stern, S., Weiser, B. & Noller, H.F. J. Mol. Biol. 204, 447–481 (1988).
Malhotra, A. & Harvey, S.C. J. Mol. Biol. 240, 308–340 (1994).
Mueller, F. & Brimacombe, R. J. Mol. Biol. 271, 524–544 (1997).
Carter, A.P. et al. Nature 407, 340–348 (2000).
Ban, N., Nissen, P., Hansen, J., Moore, P.B. & Steitz, T.A. Science 289, 905–920 (2000).
Harms, J. et al. Cell 107, 679–688 (2001).
Neefs, J., Van de Peer, Y., De Rijk, P., Chapelle, S & De Wachter, R. Nucleic Acids Res. 21, 3025–3049 (1993).
Guetell, R.R. Nucleic Acids Res. 22, 3502–3507 (1994).
Guetell, R.R. in Ribosomal RNA. Structure, Evolution, Processing, and Function in Protein Biosynthesis (eds Dahlberg, A. & Zimmerman, B.) 111–128 (CRC Press, Boca Raton; 1996).
Watson, J.D. & Crick, F.H.C. Nature 171, 737–738 (1953).
Tung, C.S. in Computation of Biomolecular Structures (eds Soumpasis, D.M. & Jobin, T.M.) 87–97 (Springer-Verlag, New York; 1993).
Tung, C.S. J. Biomol. Struct. Dyn. 17, 347–354 (1999).
Weiner, S.J., Kollman, P.A., Nguyen, D.T. & Case, D.A. J. Comput. Chem. 7, 230–252 (1986).
Pieper, U., Eswar, N., Stuart, A.C., Llyin, V.A. & Sali, A. Nucleic Acids Res. 30, 255–259 (2002).
Yusupov, M.M. et al. Science 292, 883–896 (2001).
Kozin, M.B. & Svergun, D.I. J. Appl. Crystallogr. 34, 33–41 (2001).
Gabashvili, I.S. et al. Cell 100: 537–549 (2000).
Hubbard, S.J. & Thornton, J.M. Naccess. (Department of Biochemistry and Molecular Biology, University College, London; 1993).
Nadassy, K., Wodak, S.J. & Janin, J. Biochemistry 38, 1999–2017 (1999).
Carter, A.P. et al. Science 291, 498–501 (2001).
Noller, H.F. In The RNA World (eds Gesteland, R.F., Cech, T.R. & Atkins, J.F.) 197–219 (Cold Spring Harbor Laboratory Press, New York 1999).
Yusupova, G.Z., Yusupov, M.M., Cate, J.H.D. & Noller, H.F. Cell 106, 233–241 (2001).
Benson, D.A. et al. Nucleic Acids Res. 28, 15–18 (2000).
Brosius, J., Palmer, M.L., Kennedy, P.J. & Noller, H.F. Proc. Natl. Acad. Sci. USA 75, 4801–4805 (1978).
Berman, H.M. et al. Nucleic Acids Res. 28, 235–242 (2000).
Person, W.R., Wood, T., Zhang, Z. & Miller, W. Genomics 46, 24–36 (1997).
Tatusova, T.A. & Madden, T.L FEMS Microbiol. Lett. 174, 247–250 (1999).
Wittmann, H.G. Annu. Rev. Biochem. 51, 155–183 (1982).
Makino, K. et al. Genes Genet. Syst. 74, 227–239 (1999).
Acknowledgements
This work was performed under the auspices of the Department of Energy (DOE) under contract to the University of California. C.S.T. and K.Y.S. are supported by LANL/LDRD funding. S.J. is supported by both NSF and NIH grants. The cryo-EM density map was kindly provided by J. Frank of HHMI.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Tung, CS., Joseph, S. & Sanbonmatsu, K. All-atom homology model of the Escherichia coli 30S ribosomal subunit. Nat Struct Mol Biol 9, 750–755 (2002). https://doi.org/10.1038/nsb841
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb841
This article is cited by
-
Phylogenies of the 16S rRNA gene and its hypervariable regions lack concordance with core genome phylogenies
Microbiome (2022)
-
A structural model of the E. coli PhoB Dimer in the transcription initiation complex
BMC Structural Biology (2012)
-
Spatio-temporal modeling of signaling protein recruitment to EGFR
BMC Systems Biology (2010)
-
A fast dynamic mode of the EF-G-bound ribosome
The EMBO Journal (2010)
-
Peptide nucleic acids (PNAs) antisense effect to bacterial growth and their application potentiality in biotechnology
Applied Microbiology and Biotechnology (2010)