Abstract
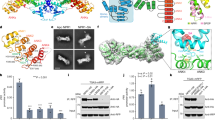
The crystal structure of p24, the single-stranded DNA (ssDNA) binding subunit of the plant defense transcription factor PBF-2, has been determined to 2.3 Å resolution. p24 is representative of a novel family of ubiquitous plant-specific proteins that we refer to as the Whirly family because of their quaternary structure. PBF-2 is composed of four p24 molecules that interact through a helix-loop-helix motif. This interaction produces a central pore, with β-strands radiating outwards, resulting in a whirligig appearance to the quaternary structure. The noncrystallographic C4 symmetry arrangement of p24 subunits is novel for ssDNA binding proteins and may explain the binding specificity of PBF-2. This structural arrangement also supports the role of PBF-2 in binding melted promoter regions to modulate gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Brivanlou, A.H. & Darnell, J.E., Jr Science 295, 813–818 (2002).
Roeder, R.G. Trends Biochem. Sci. 21, 327–335 (1996).
Kornberg, R.D. Trends Cell. Biol. 9, M46–49 (1999).
Woychik, N.A. & Hampsey, M. Cell 108, 453–463 (2002).
Nikolov, D.B. et al. Nature 360, 40–46 (1992).
Rushton, P. & Somssich, E. Curr. Opin. Plant Biol. 1, 311–315 (1998).
Maleck, K. et al. Nature Genet. 26, 403–410 (2000).
Matton, D.P. & Brisson, N. Mol. Plant Microbe Interact. 2, 325–331 (1989).
Matton, D.P., Prescott, G., Bertrand, C., Camirand, A. & Brisson, N. Plant Mol. Biol. 22, 279–291 (1993).
Després, C., Subramaniam, R., Matton, D.P. & Brisson, N. Plant Cell 7, 589–598 (1995).
Desveaux, D, Després, C., Joyeux, A., Subramaniam, R. & Brisson, N. Plant Cell 12, 1477–1489 (2000).
Hendrickson, W. & Ogata, C. Methods Enzymol. 276, 494–523 (1997).
Desveaux, D., Allard, J., Brisson, N. & Sygusch, J. Acta Crystallogr. D 58, 296–298 (2002).
Raghunathan, S., Ricard, C.S., Lohman, T.M. & Waksman, G. Proc. Natl. Acad. Sci. USA 94, 6652–6657 (1997).
Yang, C., Curth, U., Urbanke, C. & Kang, C. Nature Struct. Biol. 4, 153–157 (1997).
Rost, B. & Sander, C. J. Mol. Biol. 232, 584–599 (1993).
Rost, B. & Sander, C. Proteins 19, 55–72 (1994).
Holm, L. & Sander, C. J. Mol. Biol. 233, 123–138 (1993).
Werten, S., Wechselberger, R., Boelens, R., van der Vliet, P.C. & Kaptein, R. J. Biol. Chem. 274, 3693–3699 (1999).
Brandsen, J. et al. Nature Struct. Biol. 4, 900–903 (1997).
Bates, A.D. & Maxwell, A. In DNA Topology (eds Rickwood, D. & Male, D.) 88–95 (IRL Press, Oxford; 1993).
Duncan, R. et al. Genes Dev. 8, 465–480 (1994).
Kowalski, D., Natale, D.A. & Eddy, M.J. Proc. Natl. Acad. Sci. USA 85, 9464–9468 (1988).
Gerber, H.-P. et al. Science 263, 808–811 (1994).
Kahn, J.D., Yun, E. & Crothers, D.M. Nature 368, 163–166 (1994).
Travers, A. & Muskhelishvili, G. J. Mol. Biol. 279, 1027–1043 (1998).
Suck, D. Nature Struct. Biol. 4, 161–165 (1997).
Bochkarev, A., Pfuetzner, R.A., Edwards, A.M. & Frappier, L. Nature 385, 176–181 (1997).
Ruff, M. et al. Science 252, 1682–1689 (1991).
Klinge, C.M. Nucleic Acids Res. 29, 2905–2919 (2001).
Terwilliger, T.C. & Berendzen, J. Acta Crystallogr. D 55, 849–861 (1999).
Brünger, A.T. et al. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Acta Crystallogr. A 47, 110–119 (1991).
Diamond, R. Protein Sci. 1, 1279–1287 (1992).
Constable, C.P. & Brisson, N. Planta 188, 289–295 (1992).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. Nucleic Acids Res. 22, 4673–4680 (1994).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. J. Appl. Crystallogr. 26, 283–291 (1993).
Luzzati, P.V. Acta Crystallogr. 5, 802–810 (1952).
Acknowledgements
The assistance of T.-S. Yoon in data collection and reduction is gratefully acknowledged. Research carried out at the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences. Assistance by X8-C and X25 beamline personnel is gratefully appreciated. This research was supported by a fellowship from the Natural Sciences and Engineering Research Council of Canada (NSERC) to D.D. and research grants from the Canadian Institutes of Health Research to J.S., from NSERC to both N.B. and J.S. and the Fonds pour la Formation de Chercheurs et l'Aide à la Recherche, Québec to N.B.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Desveaux, D., Allard, J., Brisson, N. et al. A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat Struct Mol Biol 9, 512–517 (2002). https://doi.org/10.1038/nsb814
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb814
This article is cited by
-
Whirly (Why) transcription factors in tomato (Solanum lycopersicum L.): genome-wide identification and transcriptional profiling under drought and salt stresses
Molecular Biology Reports (2019)
-
Overexpression of tomato WHIRLY protein enhances tolerance to drought stress and resistance to Pseudomonas solanacearum in transgenic tobacco
Biologia plantarum (2018)
-
Identification of WHIRLY1 as a Factor Binding to the Promoter of the Stress- and Senescence-Associated Gene HvS40
Journal of Plant Growth Regulation (2014)
-
Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids
Planta (2010)
-
Recruitment of AtWHY1 and AtWHY3 by a distal element upstream of the kinesin gene AtKP1 to mediate transcriptional repression
Plant Molecular Biology (2009)