Abstract
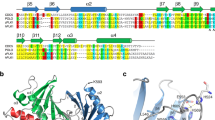
The phosphotyrosine-binding (PTB) domain of Numb, a protein involved in asymmetric cell division, has recently been shown to bind to the adapter protein Lnx through an LDNPAY sequence, to the Numb-associated kinase (Nak) through a sequence that does not contain an NPXY motif and to GP(p)Y-containing peptides obtained from library screening. We show here that these diverse peptide sequences bind with comparable affinities to the Numb PTB domain at a common binding site on the surface of the protein. The NMR structure of the Numb PTB domain in complex with a GPpY-containing peptide reveals a novel mechanism of binding with the peptide in a helical turn that does not hydrogen bond to the PTB domain β-sheet. These results suggest that PTB domains can potentially have multiple modes of peptide recognition and provide a structural basis from which the multiple functions of the Numb PTB domain during asymmetric cell division could arise.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kavanaugh, W.M. & Williams, L.T. An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science 266, 1862–1865 ( 1994).
Blaikie, P., et al. A region in Shc distinct from the SH2 domain can bind tyrosine- phosphorylated growth factor receptors. J. Biol. Chem. 269, 32031–32034 (1994).
van der Geer, P. et al. A conserved amino-terminal Shc domain binds to phosphotyrosine motifs in activated receptors and phosphopeptides. Curr. Biol. 5, 404–412 ( 1995).
Kavanaugh, W.M., Turck, C.W. & Williams, L.T. PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science 268, 1177–1179 (1995).
Trüb, T. et al. Specificity of the PTB domain of Shc for beta turn-forming pentapeptide motifs amino-terminal to phosphotyrosine. J. Biol. Chem. 270, 18205–18208 ( 1995).
Wolf, G. et al. PTB domains of Shc and IRS-1 have distinct but overlapping binding specificities. J. Biol. Chem. 270, 27407 –27510 (1995).
Borg, J.P. & Margolis, B. Function of PTB domains. Curr. Topics Microbiol. Immunol. 228, 23– 38 (1998).
Zhou, M.M. et al. Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature 378, 584– 592 (1995).
Zhou, M.M., et al. Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain. Nature Struct. Biol. 3, 388–393 (1996).
Eck, M.J., Dhe-Paganon, S., Trüb, T., Nolte, R.T. & Shoelson, S.E. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell 85, 695–705 ( 1996).
van der Geer, P. et al. Identification of residues that control specific binding of the Shc phosphotyrosine-binding domain to phosphotyrosine sites. Proc. Natl. Acad. Sci. USA 93, 963– 968 (1996).
Waksman, G. et al. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine phosphorylated peptides. Nature 358, 646–653 ( 1992).
Borg, J.P., Ooi, J., Levy, E. & Margolis, B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol. Cell. Biol. 16, 6229–6241 (1996).
Zhang, Z.T. et al. Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain. EMBO J. 16, 6141–6150 ( 1997).
Zambrano, N. et al. Interaction of the phosphotyrosine interaction/phosphotyrosine binding- related domains of Fe65 with wild-type and mutant Alzheimer's beta- amyloid precursor proteins. J. Biol. Chem. 272, 6399–6405 (1997).
Bork, P. & Margolis, B. A phosphotyrosine interaction domain. Cell 80, 693–694 (1995).
Uemura, T., Shepherd, S., Ackerman, L., Jan, L.Y. & Jan, Y.N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349–360 ( 1989).
Knoblich, J.A. Mechanisms of asymmetric cell division during animal development. Curr. Opin. Cell Biol. 9, 833–841 (1997).
Bodmer, R., Carretto, R. & Jan, Y.N. Neurogenesis of the peripheral nervous system in Drosophila embryos: DNA replication patterns and cell lineages. Neuron 3, 21–32 (1989).
Rhyu, M.S., Jan, L.Y. & Jan, Y.N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491 (1994).
Knoblich, J.A., Jan, L.Y. & Jan, Y.N. Asymmetric segregation of Numb and Prospero during cell division. Nature 377, 624– 627 (1995).
Zhong, W., Feder, J.N., Jiang, M.M., Jan, L.Y. & Jan, Y.N. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron 17, 43–53 (1996).
Verdi, J.M., et al. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 6, 1134–1145 (1996).
Zhong, W., Jiang, M.M., Weinmaster, G., Jan, L.Y. & Jan, Y.N. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development 124, 1887– 1897 (1997).
Salcini, A.E. et al. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 11, 2239–2249 (1997).
Knoblich, J.A., Jan, L.Y. & Jan, Y.N. The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc. Natl. Acad. Sci. USA 94, 13005– 13010 (1997).
Spana, E.P. & Doe, C.Q. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21–26 (1996).
Guo, M., Jan, L.Y. & Jan, Y.N. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17, 27–41 (1996).
Chien, C.T., Wang, S., Rothenberg, M., Jan, L.Y. & Jan, Y.N. Numb-associated kinase interacts with the phosphotyrosine binding domain of Numb and antagonizes the function of Numb in vivo. Mol. Cell. Biol. 18, 598–607 (1998).
Li, S.C., et al. High-affinity binding of the Drosophila Numb phosphotyrosine-binding domain to peptides containing a Gly-Pro-(p)Tyr motif. Proc. Natl. Acad. Sci. USA 94, 7204–7209 (1997).
Dho, S.E., et al. The mammalian Numb phosphotyrosine binding domain: characterization of binding specificity and identification of a novel PDZ domain-containing Numb binding protein, Lnx. J. Biol. Chem. 273, 9179–9187 (1998).
Zwahlen, C., Vincent, S.J.F., Gardner, K.H. & Kay, L.E. Significantly improved resolution for NOE correlations from valine and isoleucine (Cλ2) methyl groups in 15N,13C and 15N,13C,2H-labeled proteins. J. Am. Chem. Soc. 120, 4825–4831 (1998).
Nilges, M., Macias, M.J., O'Donoghue, S.I. & Oschkinat, H. Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin. J. Mol. Biol. 269, 408– 422 (1997).
Holm, L. & Sander, C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26, 316– 319 (1998).
Yaich, L. et al. Functional analysis of the Numb phosphotyrosine-binding domain using site-directed mutagenesis. J. Biol. Chem. 273 , 10381–10388 (1998).
Farmer, B.T., et al. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nature Struct. Biol. 3, 995– 997 (1996).
Kraut, R. & Campos-Ortega, J.A. inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeleton adaptor protein. Dev. Biol. 174, 65–81 (1996).
Kraut, R., Chia, W., Jan, L.Y., Jan, Y.N. & Knoblich, J.A. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383, 50– 55 (1996).
Kim, S.K. Polarized signaling: basolateral receptor localization in epithelial cells by PDZ-containing proteins. Curr. Opin. Cell Biol. 9, 853–859 (1997).
Neri, D., Szyperski, T., Otting, G., Senn, H. & Wüthrich, K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28, 7510 –7516 (1989).
Bax, A. Multidimensional nuclear magnetic resonance methods for protein studies. Curr. Opin. Struct. Biol. 4, 738–744 (1994).
Kay, L.E. Field gradient techniques in NMR spectroscopy. Curr. Opin. Struct. Biol. 5, 674–681 ( 1995).
Stein, E.G., Rice, L.M. & Brünger, A.T. Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J. Magn. Reson. 124, 154–164 (1997).
Rice, L.M. & Brünger, A.T. Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins: Struct. Funct. Genet. 19, 277 –290 (1994).
Brünger, A.T., et al. Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998 ).
Brünger, A.T. (Yale University, New Haven, Connecticut; 1992).
Koradi, R., Billeter, M. & Wuthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51– 5, 29–32 (1996).
Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R. & Thornton, J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–86 (1996).
Acknowledgements
We thank Y. N. Jan for kindly providing the Drosophila Numb cDNA. This work was supported by grants to J.D.F.-K. and L.E.K. from the National Cancer Institute of Canada (NCIC) and to T.P. from the Human Frontier Science Program, Asashi Chemical Company, the NCIC, and the Medical Research Council of Canada (MRC) and by Howard Hughes International Research Scholar awards to T.P. and L.E.K. T.P. is a Terry Fox Cancer Research Scientist of the NCIC. S.-C.L. is a post-doctoral fellow of the MRC. C.Z. and S.J.F.V. are recipients of Human Frontier Science Program fellowships.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, SC., Zwahlen, C., Vincent, S. et al. Structure of a Numb PTB domain–peptide complex suggests a basis for diverse binding specificity. Nat Struct Mol Biol 5, 1075–1083 (1998). https://doi.org/10.1038/4185
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/4185
This article is cited by
-
Basal condensation of Numb and Pon complex via phase transition during Drosophila neuroblast asymmetric division
Nature Communications (2018)
-
Understanding the molecular basis of substrate binding specificity of PTB domains
Scientific Reports (2016)
-
The association of cholesterol absorption gene Numb polymorphism with Coronary Artery Disease among Han Chinese and Uighur Chinese in Xinjiang, China
Lipids in Health and Disease (2015)
-
Genetic variants of numb gene were associated with elevated total cholesterol level and low density lipoprotein cholesterol level in Chinese subjects, in Xinjiang, China
Diagnostic Pathology (2015)
-
The PTB domain of ShcA couples receptor activation to the cytoskeletal regulator IQGAP1
The EMBO Journal (2010)