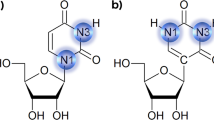
Abstract
Here we present the first solution structure of a ligand–DNA aptamer complex. Our NMR-molecular dynamics structural studies of the interaction between argininamide and a DNA stem-loop complex establishes that the hairpin loop DNA binding site undergoes an adaptive conformational transition on complex formation. The tip of the DNA loop folds down towards the stem and sandwiches the bound argininamide between reversed Hoogsteen A•C and Watson–Crick G•C base pairs. The argininamide is encapsulated within the structured DNA loop and is stabilized by an intricate set of intermolecular hydrogen bonds and stacking interactions. The structure of the complex lays out the molecular principles defining both the architecture of the internal cavity and the recognition elements that could contribute to ligand discrimination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Steitz, T.A. Structural studies of protein-nucleic acid interactions: the sources of sequence specificity. Quart. Revs. Biophys. 23, 205–280 (1990).
Joyce, G.F. In vitro evolution of nucleic acids. Curr. Opin. Struct. Biol. 4, 331–336 (1994).
Ellington, A. & Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 812–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Robertson, D.L. & Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344, 467–468 (1990).
Gold, L., Polisky, B., Uhlenbeck, O.C. & Yarus, M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 64, 763–797 (1995).
Lorsch, J.R. & Szostak, J.W. Chance and necessity in the selection of nucleic acid catalysts. Acc. Chem. Res. 29, 103–110 (1996).
Fan, P., Suri, A.K., Fiala, R., Live, D. & Patel, D.J. Molecular recognition in the FMN-RNA aptamer complex. J. Mol. Biol. 258, 480–500 (1996).
Yang, Y., Kochoyan, M., Burgstaller, P., Westhof, E. & Famulok, M. Structural basis of ligand discrimination by two related RNA aptamers revealed by NMR spectroscopy. Science 272, 1343–1347 (1996).
Jiang, F., Kumar, R.A., Jones, R.A. & Patel, D.J. Structural basis of RNA folding and recognition in an AMP-RNA aptamer complex. Nature 382, 183–186 (1996).
Dieckmann, T., Suzuki, E., Nakamura, G.K. & Feigon, J. Solution structure of an ATP-binding RNA aptamer reveals a novel fold. RNA 2, 628–640 (1996).
Ye, X., Gorin, A., Ellington, A.D. & Patel, D.J. Deep penetration of an α-helix into a widened RNA major groove in the HIV-1 rev peptide-RNA aptamer complex. Nature Struct. Biol. 3, 1026–1033 (1996).
Ellington, A.D. & Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355, 850–852 (1992).
Bock, L.C., Griffin, L.C., Latham, J.A., Vermaas, E.H. & Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355, 564–566 (1992).
Breaker, R.R. & Joyce, G.F. A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem. Biol. 2, 665–660 (1995).
Huizenga, D.E. & Szostak, J.W. A DNA aptamer that binds adenosine and ATP. Biochemistry 34, 656–665 (1995).
Li, Y., Geyer, C.R. & Sen, D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry 35, 6911–6922 (1996).
Harada, K. & Frankel, A.D. Identification of two novel arginine binding DNAs. The EMBO Journal 14, 5798–5811 (1995).
Connell, G.J., Illangesekare, M. & Yarus, M. Three small ribooligonucleotides with specific arginine sites. Biochemistry 32, 5497–5502 (1993).
Famulok, M. Molecular recognition of amino acids by RNA aptamers: An L-citrulline binding RNA motif and its evolution into an L-arginine binder. J. Am. Chem. Soc. 116, 1698–1706 (1994).
Geiger, A., Burgstaller, P., von der Eltz, H., Roeder, A. & Famulok, M. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucl. Acids Res. 24, 1029–1036 (1996).
Tao, J. & Frankel, A.D. Arginine binding RNAs resembling TAR identified by in vitro selection. Biochemistry 35, 2229–2238 (1996).
Puglisi, J.D., Chen, L., Frankel, A.D. & Williamson, J.R. Role of RNA structure in arginine recognition of TAR RNA. Proc. Natl. Acad. Sci. USA 90, 3680–3684 (1993).
Calnan, B.J., Tidor, B., Biancalana, S., Hudson, D. & Frankel, A.D. Arginine-mediated RNA recognition: the arginine fork. Science 252, 1167–1171 (1991).
Puglisi, J.D., Chen, L., Blanchard, S. & Frankel, A.D. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science 270, 1200–1203 (1995).
Ye, X., Kumar, R.A. & Patel, D.J. Molecular recognition in the bovine immunodeficiency virus tat peptide-Tar RNA complex. Chem. Biol. 2, 827–840 (1995).
Cram, D.J. & Cram, J.M. Container Molecules and their Guests. (Royal Society of Chemistry, Cambridge, UK, 1994).
Brünger, A.T. X-PLOR, A system for X-ray crystallography and NMR (Yale University Press, New Haven, CT, 1992).
Nicholls, A., Sharp, K.A. & Honig, B.H. Protein folding and association: insights from the interfacial and thermodynamics properties of hydrocarbons. Proteins 11, 281–296 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Lin, C., Patel, D. Encapsulating an amino acid in a DNA fold. Nat Struct Mol Biol 3, 1046–1050 (1996). https://doi.org/10.1038/nsb1296-1046
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb1296-1046
This article is cited by
-
Computational docking simulations of a DNA-aptamer for argininamide and related ligands
Journal of Computer-Aided Molecular Design (2015)
-
Chiral Recognition Mechanisms in Analytical Separation Sciences
Chromatographia (2012)