Abstract
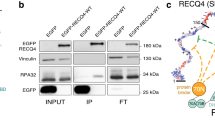
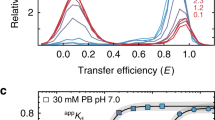
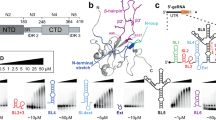
The structure of the domain from simian virus 40 (SV40) large T-antigen that binds to the SV40 origin of DNA replication (T-ag-OBD131–260) has been determined by nuclear magnetic resonance spectroscopy. The overall fold, consisting of a central five-stranded antiparallel β-sheet flanked by two α-helices on one side and one α-helix and one 310-helix on the other, is a new one. Previous mutational analyses have identified two elements, termed A (∼152–155) and B2 (203–207), as essential for origin-specific recognition. These elements form two closely juxtaposed loops that define a continuous surface on the protein. The addition of a duplex oligonucleotide containing the origin recognition pentanucleotide GAGGC induces chemical shift changes and slows amide proton exchange in resonances from this region, indicating that this surface directly contacts the DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Jacob, F., Brenner, S. & Cuzin, F. On the regulation of DNA replication in bacteria Cold Spring Harbor Sym. Quant Biol. 28, 329–348 (1963).
Stillman, B. Replicator renaissance Nature 366, 506–507 (1993).
Kornberg, A. & Baker, T.A. DNA Replication (W.H. Freeman and Co., New York, 1992).
Tegtmeyer, P. Simian virus 40 deoxyribonucleic acid synthesis: the viral replicon. J. Virol. 10, 591–598 (1972).
Borowiec, J.A., Dean, F.B., Bullock, P.A. & Hurwitz, J. Binding and unwinding-how T antigen engages the SV40 origin of DNA replication Cell 60, 181–184 (1990).
Fanning, E. & Knippers, R. Structure and function of simian virus 40 large tumor antigen. Ann. Rev. Biochem. 61, 55–85 (1992).
Brünger, A.T. X-PLOR: A System for X-ray Crystallography and NMR X-PLOR Version 3.1 Manual (Yale University Press, New Haven, CT.) (1992).
Wun-Kim, K. et al. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J. Virol. 67, 7608–7611 (1993).
Wun-Kim, K. & Simmons, D.T. Mapping of helicase and helicase substrate-binding domains on simian virus 40 large T antigen J. Virol. 64, 2014–2020 (1990).
Simmons, D.T., Loeber, G. & Tegtmeyer, P. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J. Virol. 64, 1973–1983 (1990).
Simmons, D.T., Wun-Kim, K. & Young, W. Identification of simian virus 40 T-antigen residues important for specific and nonspecific binding to DNA and for helicase activity. J. Virol. 64, 4858–4865 (1990).
Tjian, R. The binding site on SV40 DNA for a T-antigen related protein. Cell 13, 165–179 (1978).
Bax, A., Ikura, M., Kay, I.E., Torchia, D.A. & Tschudin, R. Comparison of different modes of two-dimensional reverse-correlation NMR for the study of proteins J. Magn. Reson. 86, 304–318 (1990).
Lian, L.Y., Barsukov, I.L., Sutcliffe, M.J., Sze, K.H. & Roberts, G.C.K. Protein-Ligand Interactions: Exchange Processes and Determination of Ligand Conformation and Protein-Ligand Contacts Meth. Enzym. 239, 657–700 (1994).
Cleland, W.W. & Kreevoy, M.M. Low-barrier hydrogen bonds and enzymatic catalysis Science 264, 1887–1890 (1994).
Frey, P.A., Whitt, S.A. & Tobin, J.B. A low-barrier hydrogen bond in the catalytic triad of serine proteases Science 264, 1927–1930 (1994).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 170, 123–138 (1993).
Bernstein, F.C. et al. The protein data bank: a computer-based archival file for macromolecular structures J. Mol. Biol. 112, 535–542 (1977).
Abola, E.E., Bernstein, F.C., Bryant, S.H., Koetzle, T.F. & Weng, J. “Protein Data Bank” in crystallographic databases-information content, software systems, scientific applications Data commission of the Int'l union of crystallography, Bonn/Cambridge/Chester 107–132 (1987).
Avis, J.M. et al. Solution structure of the N-terminal RNP domain of U1A protein: the role of the C-terminal residues in structure stability and RNA binding. J. Mol. Biol. 257, 398–411 (1996).
Hegde, R.S., Grossman, S.R., Laimins, L.A. & Sigler, P.B. Crystal structure at 1.7 Å of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature 359, 505–512 (1992).
Bochkarev, A. et al. Crystal structure of the DNA-binding domain of the Epstein-barr virus origin-binding protein, EBNA1, bound to DNA Cell 84, 791–800 (1996).
Harrison, S.C. A structural taxonomy of DNA-binding domains Nature 353, 715–719 (1991).
Kay, L.E., Ikura, M., Tschudin, R. & Bax, A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins J. Magn. Reson. 89, 496–514 (1990).
Bax, A. & Ikura, M. An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the alpha-carbon of the preceding residue in uniformly 15N/13C enriched proteins J. Biomolec. NMR 1, 99–104 (1991).
Farmer II, B.T., Venters, R.A., Spicer, L.D., Wittekind, M.G. & Müller, L. A refocused and optimized HNCA: increased sensitivity and resolution in large macromolecules. J. Biomolec. NMR 2, 195–202 (1992).
Farmer II, B.T. Optimized triple resonance applied to peptides labeled only with 15N. The HN(CO)(CA) experiment J. Magn. Reson. 94, 413–418 (1991).
Kay, L.E. & Bax, A. New methods for the measurement of NH-CαH coupling constants in 15N-labeled proteins J. Magn. Reson. 86, 110–126 (1990).
Vuister, G.W. & Bax, A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNHα) coupling constants in 15N-enriched proteins J. Am. Chem. Soc. 115, 7772–7777 (1993).
Carson, M. Ribbon models of macromolecules J. Mol. Graphics 5, 103–106 (1987).
Nicholls, A.J., Sharp, K.A. & Honig, B. Protein folding and association: insights from interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Luo, X., Sanford, D., Bullock, P. et al. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat Struct Mol Biol 3, 1034–1039 (1996). https://doi.org/10.1038/nsb1296-1034
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb1296-1034
This article is cited by
-
Mechanism of DNA translocation in a replicative hexameric helicase
Nature (2006)
-
Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex
The EMBO Journal (2006)
-
Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA
The EMBO Journal (2006)
-
Insights into hRPA32 C-terminal domain–mediated assembly of the simian virus 40 replisome
Nature Structural & Molecular Biology (2005)
-
Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen
Nature (2003)