Abstract
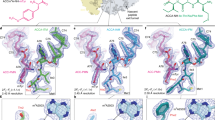
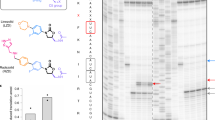
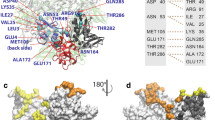
The cytotoxin sarcin disrupts elongation factor binding and protein synthesis by specifically cleaving one phosphodiester bond in ribosomes. To elucidate the molecular basis of toxin action, we determined three cocrystal structures of the sarcin homolog restrictocin bound to different analogs that mimic the target sarcin/ricin loop (SRL) structure of the rat 28S rRNA. In these structures, restrictocin contacts the bulged-G motif and an unfolded form of the tetraloop of the SRL RNA. In one structure, toxin loops guide selection of the target site by contacting the base critical for recognition (G4319) and the surrounding S-shaped backbone. In another structure, base flipping of the tetraloop enables cleavage by placing the target nucleotide in the active site with the nucleophile nearly inline for attack on the scissile bond. These structures provide the first views of how a site-specific protein endonuclease recognizes and cleaves a folded RNA substrate.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Steyaert, J. Eur. J. Biochem. 247, 1–11 (1997).
Arni, R.K. et al. Biochemistry 38, 2452–2461 (1999).
Wool, I.G. In Ribonucleases structures and function (eds D'Alessio, G. & Riordan, J.F.) 131–162 (Academic Press, Inc., San Diego; 1997).
Olmo, N. et al. Eur. J. Biochem. 268, 2113–2123 (2001).
Yang, X. & Moffat, K. Structure 4, 837–852 (1996).
Lacadena, J. et al. Proteins 37, 474–484 (1999).
Kao, R. & Davies, J. J. Biol. Chem. 274, 12576–12582 (1999).
Munishkin, A. & Wool, I.G. Proc. Natl. Acad. Sci. USA 94, 12280–12284 (1997).
Ban, N., Nissen, P., Hansen, J., Moore, P.B. & Steitz, T.A. Science 289, 905–920 (2000).
Moore, P.B. Annu. Rev. Biochem. 68, 287–300 (1999).
Correll, C.C. et al. Proc. Natl. Acad. Sci. USA 95, 13436–13441 (1998).
Moazed, D., Robertson, J.M. & Noller, H.F. Nature 334, 362–364 (1988).
Takeda, E., Bi, X., Yoshinari, S. & Endo, Y. Nucleic Acids Symp. 37, 131–132 (1997).
Gluck, A., Endo, Y. & Wool, I.G. Nucleic Acids Res. 22, 321–324 (1994).
Correll, C.C., Wool, I.G. & Munishkin, A. J. Mol. Biol. 292, 275–287 (1999).
Antao, V.P., Lai, S.Y. & Tinoco, I. Nucleic Acids Res. 19, 5901–5905 (1991).
Endo, Y., Gluck, A., Chan, Y.L., Tsurugi, K. & Wool, I.G. J. Biol. Chem. 265, 2216–2222 (1990).
Soukup, G.A. & Breaker, R.R. RNA 5, 1308–1325 (1999).
Diener, J.L. & Moore, P.B. Mol. Cell 1, 883–894 (1998).
Scott, W.G., Finch, J.T. & Klug, A. Cell 81, 991–1002 (1995).
Cai, Z. & Tinoco, I. Biochemistry 35, 6026–6036 (1996).
Wedekind, J.E. & McKay, D.B. Nature Struct. Biol. 6, 261–268 (1999).
Rupert, P.B. & Ferre-D'Amare, A.R. Nature 410, 780–786 (2001).
Richards, F.M. et al. Cold Spring Harbor Symp. Quant. Biol. 36, 35–43 (1971).
Li, H., Trotta, C.R. & Abelson, J. Science 280, 279–284 (1998).
Weston, S.A., Tucker, A.D., Thatcher, D.R., Derbyshire, D.J. & Pauptit, R.A. J. Mol. Biol. 244, 410–422 (1994).
Pingoud, A. & Jeltsch, A. Eur. J. Biochem. 246, 1–22 (1997).
Roberts, R.J. & Cheng, X. Annu. Rev. Biochem. 67, 181–198 (1998).
Otwinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 (1997).
Navaza, J. Acta Crystallogr. A 50, 157–163 (1994).
Read, R.J. Acta Crstallolgr. A 42, 140–149 (1986).
Cowtan, K.D. & Zhang, K.Y. Prog. Biophys. Mol. Biol. 72, 245–270 (1999).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard . Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Acta Crystallogr. D 54, 905–921 (1998).
Carson, M. Methods Enzymol. 277, 493–505 (1991).
Kraulis, P.J. J. Appl. Crystallogr. 24, 946–950 (1991).
Seggerson, K. & Moore, P.B. RNA 4, 1203–1215 (1998).
Wimberly, B.T., Guymon, R., McCutcheon, J.P., White, S.W. & Ramakrishnan, V. Cell 97, 491–502. (1999).
Acknowledgements
We thank J. Beneken, Y. Chen, A. Glück, T. Pan, P. Rice and I. Wool for helpful discussion and the staff of BioCARS for help with data collection. A Cancer Research Foundation Young Investigator Award and grant from the NIH to C.C.C. supported this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, X., Gérczei, T., Glover, L. et al. Crystal structures of restrictocin–inhibitor complexes with implications for RNA recognition and base flipping. Nat Struct Mol Biol 8, 968–973 (2001). https://doi.org/10.1038/nsb1101-968
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb1101-968
This article is cited by
-
High-affinity recognition of specific tRNAs by an mRNA anticodon-binding groove
Nature Structural & Molecular Biology (2019)
-
RNAfitme: a webserver for modeling nucleobase and nucleoside residue conformation in fixed-backbone RNA structures
BMC Bioinformatics (2018)
-
De novo computational RNA modeling into cryo-EM maps of large ribonucleoprotein complexes
Nature Methods (2018)
-
Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity
Nature Structural & Molecular Biology (2016)
-
Atomistic details of the molecular recognition of DNA-RNA hybrid duplex by ribonuclease H enzyme
Journal of Chemical Sciences (2015)