Abstract
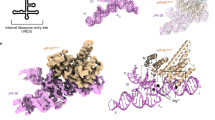
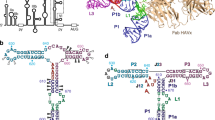
Complex RNA structures regulate many biological processes, but are often too large for structure determination by NMR methods. The 5′ untranslated region (5′ UTR) of the hepatitis C viral (HCV) RNA genome contains an internal ribosome entry site (IRES) that binds to 40S ribosomal subunits with high affinity and specificity to control translation. Domain II of the HCV IRES forms a 25-kDa folded subdomain that may alter ribosome conformation. We report here the structure of domain II as determined using an NMR approach that combines short- and long-range structural data. Domain II adopts a distorted L-shape structure, and its overall shape in the free form is markedly similar to its 40S subunit–bound form; this suggests how domain II may modulate 40S subunit conformation. The results show how NMR can be used for structural analysis of large biological RNAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pestova, T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J. & Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12, 67–83 (1998).
Sachs, A.B., Sarnow, P. & Hentze, M.W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89, 831–838 (1997).
Kieft, J.S., Zhou, K., Jubin, R. & Doudna, J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7, 194–206 (2001).
Spahn, C.M. et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291, 1959–1962 (2001).
Otto, G.A., Lukavsky, P.J., Lancaster, A.M., Sarnow, P. & Puglisi, J.D. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA 8, 913–923 (2002).
Kim, I., Lukavsky, P.J. & Puglisi, J.D. NMR study of 100kDa HCV IRES RNA using segmental isotope labeling. J. Am. Chem. Soc. 124, 9338–9339 (2002).
Tjandra, N. & Bax, A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science 278, 1111–1114 (1997).
Clore, G.M., Gronenborn, A.M. & Tjandra, N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 131, 159–162 (1998).
McCallum, S.A. & Pardi, A. Refined solution structure of the iron-responsive element RNA using residual dipolar couplings. J. Mol. Biol. 326, 1037–1050 (2003).
Honda, M., Beard, M.R., Ping, L.H. & Lemon, S.M. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73, 1165–1174 (1999).
Zhao, W.D. & Wimmer, E. Genetic analysis of a poliovirus/hepatitis C virus chimera: new structure for domain II of the internal ribosomal entry site of hepatitis C virus. J. Virol. 75, 3719–3730 (2001).
Gonzalez, R.L., Jr., & Tinoco I., Jr. Identification and characterization of metal ion binding sites in RNA. Methods Enzymol. 338, 421–443 (2001).
Lukavsky, P.J., Otto, G.A., Lancaster, A.M., Sarnow, P. & Puglisi, J.D. Structures of two essential RNA domains for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 7, 1105–1110 (2000).
Correll, C.C. et al. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl. Acad. Sci. USA 95, 13436–13441 (1998).
Smith, D.B. et al. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. The International HCV Collaborative Study Group. J. Gen. Virol. 76, 1749–1761 (1995).
Puglisi, J.D. & Wyatt, J.R. Biochemical and NMR studies of RNA conformation with an emphasis on RNA pseudoknots. Methods Enzymol. 261, 323–350 (1995).
Lukavsky, P.J. & Puglisi, J.D. RNAPack: an integrated NMR approach to RNA structure determination. Methods 25, 316–332 (2001).
Batey, R.T., Inada, M., Kujawinski, E., Puglisi, J.D. & Williamson, J.R. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res. 20, 4515–4523 (1992).
Hansen, M.R., Mueller, L. & Pardi, A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 5, 1065–1074 (1998).
Goddard, T.D. & Kneller, D.G. Sparky 3. (University of California, San Francisco, 2000)
Dingley, A.J. & Grzesiek, S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 120, 8293–8297 (1998).
Weigelt, J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J. Am. Chem. Soc. 120, 10778–10779 (1998).
Pervushin, K., Riek, R., Wider, G. & Wüthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 94, 12366–12371 (1997).
Brunger, A.T. X-PLOR, Version 3.1. A System for X-ray Crystallography and NMR (Yale Univ. Press, New Haven, Connecticut, USA, 1992).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Losonczi, J.A., Andrec, M., Fischer, M.W. & Prestegard, J.H. Order matrix analysis of residual dipolar couplings using singular value decomposition. J. Magn. Reson. 138, 334–342 (1999).
Zweckstetter, M. & Bax, A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J. Am. Chem. Soc. 122, 3791–3792 (2000).
Lavery, R. & Sklenar, H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 6, 655–667 (1989).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 (1996).
Acknowledgements
We thank E. Lau for labeled nucleotide preparation and P. Sarnow, Y. Shibata-Lukavsky, R.L. Gonzalez, D. Daniels, C. Liu and E.V. Puglisi for helpful discussions. This work was supported by grants from the US National Institutes of Health, the Hutchison Foundation and Eli Lilly. Stanford Magnetic Resonance Laboratory is supported by the Stanford School of Medicine.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Lukavsky, P., Kim, I., Otto, G. et al. Structure of HCV IRES domain II determined by NMR. Nat Struct Mol Biol 10, 1033–1038 (2003). https://doi.org/10.1038/nsb1004
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb1004
This article is cited by
-
Systematic detection of tertiary structural modules in large RNAs and RNP interfaces by Tb-seq
Nature Communications (2023)
-
RNA nanostructure transformation into DNA ones
Nano Research (2022)
-
Structural-profiling of low molecular weight RNAs by nanopore trapping/translocation using Mycobacterium smegmatis porin A
Nature Communications (2021)
-
QRNAS: software tool for refinement of nucleic acid structures
BMC Structural Biology (2019)
-
Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution
Nature Communications (2015)