Abstract
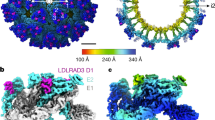
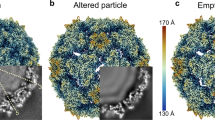
Group B coxsackieviruses (CVB) utilize the coxsackievirus-adenovirus receptor (CAR) to recognize host cells. CAR is a membrane protein with two Ig-like extracellular domains (D1 and D2), a transmembrane domain and a cytoplasmic domain. The three-dimensional structure of coxsackievirus B3 (CVB3) in complex with full length human CAR and also with the D1D2 fragment of CAR were determined to ∼22 Å resolution using cryo-electron microscopy (cryo-EM). Pairs of transmembrane domains of CAR associate with each other in a detergent cloud that mimics a cellular plasma membrane. This is the first view of a virus–receptor interaction at this resolution that includes the transmembrane and cytoplasmic portion of the receptor. CAR binds with the distal end of domain D1 in the canyon of CVB3, similar to how other receptor molecules bind to entero- and rhinoviruses. The previously described interface of CAR with the adenovirus knob protein utilizes a side surface of D1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rueckert, R.R. In Fields virology (eds Fields, B.N., D.M. Knipe & P.M. Howley) 609–654 (Lippincott-Raven Press, Philadelphia & New York; 1996).
Melnick, J.L. In Fields virology (eds Fields, B.N., D.M. Knipe & P.M. Howley) 655–712 (Lippincott-Raven Publishers, Philadelphia & New York; 1996).
Muckelbauer, J.K. et al. Structure 3, 653–668 (1995).
Lonberg-Holm, K., Crowell, R.L. & Philipson, L. Nature 259, 679–681 (1976).
Bergelson, J.M. et al. Science 275, 1320–1323 (1997).
Wang, X. & Bergelson, J.M. J. Virol. 73, 2559–2562 (1999).
Martino, T.A. et al. Virology 271, 99–108 (2000).
Rossmann, M.G. et al. Nature 317, 145–153 (1985).
Kolatkar, P.R. et al. EMBO J. 18, 6249–6259 (1999).
Olson, N.H. et al. Proc. Natl. Acad. Sci. USA 90, 507–511 (1993).
He, Y. et al. Proc. Natl. Acad. Sci. USA 97, 79–84 (2000).
Belnap, D.M. et al. Proc. Natl. Acad. Sci. USA 97, 73–78 (2000).
Xing, L. et al. EMBO J. 19, 1207–1216 (2000).
Xiao, C. et al. J. Virol. 75, 2444–2451 (2001).
Rossmann, M.G. Protein Sci. 3, 1712–1725 (1994).
Oliveira, M.A. et al. Structure 1, 51–68 (1993).
Smith, T.J., Chase, E.S., Schmidt, T.J., Olson, N.H. & Baker, T.S. Nature 383, 350–354 (1996).
Tomko, R.P., Xu, R. & Philipson, L. Proc. Natl. Acad. Sci. USA 94, 3352–3356 (1997).
Nemerow, G.R. Virology 274, 1–4 (2000).
Bai, M., Harfe, B. & Freimuth, P. J. Virol. 67, 5198–5205 (1993).
Bewley, M., Springer, K., Zhang, Y.B., Freimuth, P. & Flanagan, J.M. Science 286, 1579–1583 (1999).
Knowlton, K.U., Jeon, E.S., Berkley, N., Wessely, R. & Huber, S. J. Virol. 70, 7811–7818 (1996).
Bergelson, J.M. et al. J. Virol. 72, 415–419 (1998).
van Raaij, M.J., Chouin, E., van der Zandt, H., Bergelson, J.M. & Cusack, S. Structure 8, 1147–1155 (2000).
Gauntt, C.J., Trousdale, M.D., LaBadie, D.R., Paque, R.E. & Nealon, T. J. Med. Virol. 3, 207–220 (1979).
Mayr, G.A. & Freimuth, P. J. Virol. 71, 412–418 (1997).
Freimuth, P. et al. J. Virol. 73, 1392–1398 (1999).
Robison, C.S. & Whitt, M.A. J. Virol. 74, 2239–2246 (2000).
Baker, T.S., Olson, N.H. & Fuller, S.D. Microbiol. Mol. Biol. Rev. 63, 862–922 (1999).
Baker, T.S. & Cheng, R.H. J. Struct. Biol. 116, 120–130 (1996).
Freigang, J. et al. Cell 101, 425–433 (2000).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Acta Crystallogr. A 47, 110–119 (1991).
Rossmann, M.G. Acta Crystallogr. D 56, 1341–1349 (2000).
Acknowledgements
We thank J. DeGregori at the University of Colorado Cancer Center for the hybridoma cells that we initially used to produce CAR. We thank the BNL genome sequencing group, especially B. Lade, L. Butler, K. Pellechi, J. Kieleczawa and J. Dunn, for sequence analysis of the BAC containing the CAR gene. DNA sequencing was supported in part by the Office of Biological and Environmental Research of the U.S. Department of Energy. We also thank C. Xiao, W. Zhang, S. Mukhopadhyay, B. Hébert and J. Henderson for helpful discussions, C. Towell and S. Wilder for help in preparation of the manuscript, and K. Springer for purification of the CAR D1D2 protein fragment. This work was supported by NIH grants to M.G.R., R.J.K., P.F., M.A.W. and T.S.B., and grants from the Keck Foundation and Purdue University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
He, Y., Chipman, P., Howitt, J. et al. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat Struct Mol Biol 8, 874–878 (2001). https://doi.org/10.1038/nsb1001-874
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb1001-874
This article is cited by
-
Human Coxsackie- and adenovirus receptor is a putative target of neutrophil elastase-mediated shedding
Molecular Biology Reports (2022)
-
Myocarditis and inflammatory cardiomyopathy: current evidence and future directions
Nature Reviews Cardiology (2021)
-
Cellular receptors for enterovirus A71
Journal of Biomedical Science (2020)
-
Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage
Nature Communications (2020)
-
Cardiovascular Complications in Patients with COVID-19: Consequences of Viral Toxicities and Host Immune Response
Current Cardiology Reports (2020)