Abstract
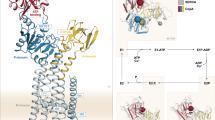
Cellular systems for handling transition metal ions have been identified, but little is known about the structure and function of the specific trafficking proteins. The 1.8 Å resolution structure of the yeast copper chaperone for superoxide dismutase (yCCS) reveals a protein composed of two domains. The N-terminal domain is very similar to the metallochaperone protein Atx1 and is likely to play a role in copper delivery and/or uptake. The second domain resembles the physiological target of yCCS, superoxide dismutase I (SOD1), in overall fold, but lacks all of the structural elements involved in catalysis. In the crystal, two SOD1-like domains interact to form a dimer. The subunit interface is remarkably similar to that in SOD1, suggesting a structural basis for target recognition by this metallochaperone.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Culotta, V.C. et al. J. Biol. Chem. 272, 23469– 23472 (1997).
Pufahl, R.A. et al. Science 278, 853– 856 (1997).
Valentine, J.S. & Gralla, E.B. Science 278, 817–818 (1997).
Rae, T.D., Schmidt, P.J., Pufahl, R.A., Culotta, V.C. & O'Halloran, T.V. Science 284, 805–808 (1999).
Lin, S.-J., Pufahl, R.A., Dancis, A., O'Halloran, T.V. & Culotta, V.C. J. Biol. Chem. 272, 9215– 9220 (1997).
Gamonet, F. & Lauquin, G.J. Eur. J. Biochem. 251 , 716–723 (1998).
Glerum, D.M., Shtanko, A. & Tzagoloff, A. J. Biol. Chem. 271, 14504– 14509 (1996).
Srinivasan, C., Posewitz, M.C., George, G.N. & Winge, D.R. Biochemistry 37, 7572–7577 (1998).
Rosenzweig, A.C. et al. Structure 7, 605–617 (1999).
Gitschier, J., Moffat, B., Reilly, D., Wood, W.I. & Fairbrother, W.J. Nature Struct. Biol. 5, 47– 54 (1998).
Steele, R.A. & Opella, S.J. Biochemistry 36, 6885–6895 (1997).
Bull, P.C. & Cox, D.W. Trends Genet. 10, 246–252 (1994).
Kissinger, C.R., Sieker, L.C., Adman, E.T. & Jensen, L.H. J. Mol. Biol. 219, 693–715 (1993).
Portnoy, M.E. et al. J. Biol. Chem. 274, 15041– 15045 (1999).
Lyons, T.J. et al. J. Biol. Inorg. Chem. 3, 650– 662 (1998).
Tainer, J.A., Getzoff, E.D., Beem, K.M., Richardson, J.S. & Richardson, D.C. J. Mol. Biol. 160 , 181–217 (1982).
Getzoff, E.D., Tainer, J.D., Stempien, M.M., Bell, G.I. & Hallewell, R.A. Proteins 5, 322–336 (1989).
Bordo, D., Djinovic, K. & Bolognesi, M. J. Mol. Biol. 238, 366– 386 (1994).
Getzoff, E.D. et al. Nature 306, 287–290 (1983).
Djinovic, K. et al. J. Mol. Biol. 225, 791– 809 (1992).
Jones, S. & Thornton, J.M. Proc. Natl. Acad. Sci. USA 93, 13–20 (1996).
Bertini, I., Piccioli, M., Viezzoli, M.S., Chiu, C.Y. & Mullenbach, G.T. Eur. J. Biophys. 23, 167–176 (1994).
Stites, W.E. Chem. Rev. 97, 1233–1250 (1997).
Valentine, J.S. & Pantoliano, M.W. in Copper proteins (ed. Spiro, T.G.) 291–358 (Wiley-Interscience, New York; 1981).
Siddique, T., Nijhawan, D. & Hentati, A. J. Neural Transm. 49, 219– 233 (1997).
Hendrickson, W.A., Horton, J.R. & LeMaster, D.M. EMBO J. 9, 1665– 1672 (1990).
Otwinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 ( 1997).
Collaborative Computational Project, Number 4 Acta Crystallogr. D50, 760–763 (1994).
Brünger, A.T. et al. Acta Crystallogr. D54, 905– 921 (1998).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Acta Crystallogr. A47, 110–119 ( 1991).
Laskowski, R.A. J. Appl. Crystallogr. 26, 283–291 (1993).
Kraulis, P.J. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E.A. & Bacon, D.J. Methods Enzymol. 277 , 505–524 (1997).
Acknowledgements
We thank L. Pascoli and M. Hou for assistance with crystallization and J. Quintana and D. Keane for assistance with data collection. This work was supported by a grant from the NIH (to A.C.R.), by funds from the ALS Association (A.C.R.), by funds from the Robert H. Lurie Cancer Center (A.C.R.), by a grant from the NIH (to T.V.O.), by a supplement from NIGMS to this same grant (to A.C.R. and T.V.O.), by funds from the ALS Association (T.V.O.), and by an NIH NRSA Training Grant (R.A.P.). The DND-CAT Synchrotron Research Center at the Advanced Photon Sourceis supported by the E.I. Dupont de Nemours & Co., The Dow Chemical Company, the NSF and the State of Illinois.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lamb, A., Wernimont, A., Pufahl, R. et al. Crystal structure of the copper chaperone for superoxide dismutase. Nat Struct Mol Biol 6, 724–729 (1999). https://doi.org/10.1038/11489
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/11489
This article is cited by
-
The essential liaison of two copper proteins: the Cu-sensing transcription factor Mac1 and the Cu/Zn superoxide dismutase Sod1 in Saccharomyces cerevisiae
Current Genetics (2023)
-
Soluble and membrane-bound protein carrier mediate direct copper transport to the ethylene receptor family
Scientific Reports (2019)
-
Exploring the Extended Biological Functions of the Human Copper Chaperone of Superoxide Dismutase 1
The Protein Journal (2019)
-
Copper–zinc superoxide dismutase (Sod1) activation terminates interaction between its copper chaperone (Ccs) and the cytosolic metal-binding domain of the copper importer Ctr1
BioMetals (2019)
-
Inhibition of human copper trafficking by a small molecule significantly attenuates cancer cell proliferation
Nature Chemistry (2015)