Abstract
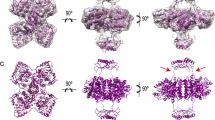
Tyrosine hydroxylase (TyrOH) catalyzes the conversion of tyrosine to L-DOPA, the rate-limiting step in the biosynthesis of the catecholamines dopamine, adrenaline, and noradrenaline. TyrOH is highly homologous in terms of both protein sequence and catalytic mechanism to phenylalanine hydroxylase (PheOH) and tryptophan hydroxylase (TrpOH). The crystal structure of the catalytic and tetramerization domains of TyrOH reveals a novel α-helical basket holding the catalytic iron and a 40 Å long anti-parallel coiled coil which forms the core of the tetramer. The catalytic iron is located 10 Å below the enzyme surface in a 17 Å deep active site pocket and is coordinated by the conserved residues His 331, His 336 and Glu 376. The structure provides a rationale for the effect of point mutations in TyrOH that cause L-DOPA responsive parkinsonism and Segawa's syndrome. The location of 112 different point mutations in PheOH that lead to phenylketonuria (PKU) are predicted based on the TyrOH structure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Mallet, J. The TIPS/TINS lecture — Catecholamines — from gene regulation to neuropsychiatric disorders. Trends Neurosci. 19, 191–196 (1996).
Kaufman, S. Studies on the mechanism of the enzymatic conversion of phenylalanine to tyrosine. J. Biol. Chem. 234, 2677–2682 (1959).
Smyth, C. et al. Further tests for linkage of bipolar affective disorder to the tyrosine hydroxylase gene locus on chromosome 11p15 in a new series of multiplex British affective disorder pedigrees. Amer. J. of Psych. 153, 271–274 (1996).
Thibaut, F. et al. Association of DNA polymorphism in the first intron of the tyrosine hydroxylase gene with disturbances of the catecholaminergic system in schizophrenia. Schizophrenia Res. 23, 259–264 (1997).
Ludecke, B. et al. Recessively inherited L-DOPA responsive parkinsonism in infancy caused by a point mutation (L205P) in the tyrosine hydroxylase gene. Hum. Mol. Gen. 7, 1023–1028 (1996).
Ludecke, B., Dworniczak, B., Bartholome, K. A point mutation in the tyrosine hydroxylase gene associated with Segawa's syndrome. Hum. Genet. 95, 123–125 (1995).
Fitzpatrick, P.F. Steady-state kinetic mechanism of rat tyrosine hydroxylase. Biochemistry 30, 3658–3662 (1991).
Fitzpatrick, P.F. Kinetic isotope effects on hydroxylation of ring-deuterated phenylalanines by tyrosine hydroyxlase provide evidence against partitioning of an arene oxide intermediate. J. Am. Chem. Soc. 116, 1133–1134 (1994).
Hillas, P.J. and Fitzpatrick, P.F. A mechanism for hydroxylation by tyrosine hydroxylase based on partitioning of substituted phenylalanines. Biochemistry 35, 6969–6975 (1996).
Hufton, S.E., Jennings, I.G., Cotton, R.G.H. Structure and function of the aromatic amino acid hydroxylases. Biochem. J. 311, 353–366 (1995).
Fitzpatrick, P.F. Studies of the rate-limiting step in the tyrosine hydroxylase reaction: alternate substrates, solvent isotope effects, and transition-state analogues. Biochemistry 30, 6386–6391 (1991).
Daubner, S.C., Lohse, D.L., Fitzpatrick, P.F. Expression and characterization of the catalytic and regulatory domains of rat tyrosine hydroxylase. Prot. Sci. 2, 1452–1460 (1993).
Rost, B. PHD: Predicting one-dimensional protein structure by profile-based neural networks. Meth.Enz. 226, 525–539 (1996).
Dickson, P.W., Jennings, I.G., Cotton, R.G.H. Delineation of the catalytic core of phenylalanine hydroxylase and identification of glutamate 286 as a critical residue for pterin function. J. Bio. Chem. 269, 20369–20375 (1994).
Vrana, K.E., Walker, S.J., Rucker, P., Liu, X. A carboxyl terminal leucine zipper is required for tyrosine hydroxylase tetramer formation. J. Neurochem. 63, 2014–2020 (1994).
Lohse, D.L., Fitzpatrick, P.F. Identification of the intersubunit binding region in rat tyrosine hydroxylase. Biochem. Biophys. Res. Com. 197, 1543–1548 (1993).
Lupas, A. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21, 375–382 (1996).
Bennett, M.J., Schlunegger, M.P., Eisenberg, D. 3D domain swapping — A mechanism for oligomer assembly. Prot. Sci. 4, 2455–2468 (1995).
Quinsey, N.S., Lenaghan, C.M., Dickson, P.W. Identification of Gln(313) and Pro(327) as residues critical for substrate inhibition in tyrosine hydroxylase. J. Neurochem. 66, 908–914 (1996).
The FSSP database [ Holm, L., Ouzounis, C., Sander, C., Tuparev, G., Vriend, G. Protein Sci. 1, 1691–1698 (1992); Holm, L. and Sander, C. Nucleic Adds Res. 22, 3600–3609 (1994)] is a compilation of representative 3-D protein structures based on an automated analysis of the Protein Data Bank [ Bernstein, F.C. et al., J. Mol. Biol. 233, 123 (1993)].
Iwaki, M., Phillips, R.S., Kaufman, S. Proteolytic modification of the amino-terminal and carboxyl-terminal regions of rat hepatic phenylalanine hydroxylase. 261, 2051–2056 (1986).
Ramsey, A.J., Daubner, S.C., Ehrlich, J.I., Fitzpatrick, P.F. Identification of iron ligands in tyrosine hydroxylase by mutagenesis of conserved histidine residues. Prot. Sci. 4, 2082–2086 (1985).
Meyerklaucke, W. et al.. Mossbauer, electron-paramagnetic-resonance and X-ray absorption fine-structure studies of the iron environment in recombinant tyrosine hydroxylase. Eur. J. Biochem. 241, 432–439 (1996).
Ramsey, A.J., Hillas, P.J., Fitzpatrick, P.F. Characterization of the active site iron in tyrosine hydroxylase — redox states of the iron. J. Bio. Chem. 271, 24395–24400 (1996).
Han, S., Eltis, L.D., Timmis, K.N., Muchmore, S.W., Bolin, J.T. Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science 270, 976–980 (1995).
Senda, T. et al. Three-dimensional structures of free form and two substrate complexes of an extradiol ring-cleavage type dioxygenase, the BPHC enzyme from pseudomonas SP strain KKS102. J. Mol. Biol. 255, 735–752 (1996).
Quinsey, N.S., Lenaghan, C.M, Dickson, P.W. Identification of Gln(313) and Pro(327) as residues critical for substrate inhibition. J. Neurochem. 66, 908–914 (1996).
Knappskogg, P.M., Flatmark, T., Mallet, J., Ludecke, B., Bartholome, K. Recessively inherited L-DOPA-responsive dystonia caused by a point mutation (Q381K) in the tyrosine hydroxylase gene. Hum. Mol. Gen. 4, 1209–1212 (1995).
Eisensmith, R.C., Woo, S.L.C. Molecular genetics of PKU: from molecular anthropology to gene therapy. Adv. Genetics 32, 199–271 (1995).
Hendrickson, W.A., Horton, J.R., LeMaster, D.M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD) — A vehicle for direct determination of 3-dimensional structure. EMBO J. 9, 1665–1672 (1990).
Otwinowski, Z. in Data Collection and Processing (eds L. Sawyer, N. Isaacs, S. Bailey) 56562 (Science and Engineering Research Council, Warrington, UK; 1993).
CCP4: A Suite of Programs for Protein Crystallography (SERC Daresbury Laboratory, Warrington WA44AD, UK, 1979).
McRee, D.E. A visual protein crystallographic software system for X11/Xview J. Mol. Graphics 10, 44 (1992).
Otwinowski, Z. in Proceedings of the CCP4 Study Weekend: Isomorphous Replacement and Anomalous Scattering (eds W. Wolf, P.R. Evans, A.G.W. Leslie) 80886 (SERC Daresbury Laboratory, Warrington, UK; 1991).
Cowtan, K.D. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr. D49, 148–157 (1993).
Jones, T.A., Zou, J.-Y., Cowan, S.W., KKjelgaard, M. Improved methods for the building of protein models in electron density maps and the location of errors in these. Acta Crystallogr. A47, 110–119 (1991).
Brünger, A.T. X-PLOR v3.8, Yale Univ. Press, New Haven, CT (1996).
Colovos, C., Yeates, T.O. Verification of protein structures — Patterns of nonbonded atomic interactions. Prot. Sci. 2, 1511–1519 (1993).
Laskowski, R.A., MacArthus, M.W., Moss, D.S., Thornton, J.M. PROCHECK — A program to check the stereochemical quality of protein structures. J. Applied Crystallogr. 26, 283–291 (1993).
Vriend, G. What If: A molecular modeling and drug design program. J. Mol. Graphics 8, 52–56 (1990).
Kabsch, W., Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Kraulis, P.J. MOLSCRIPT — A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E.A., Murphy, M.E.P. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D50, 869–873 (1994).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Goodwill, K., Sabatier, C., Marks, C. et al. Crystal structure of tyrosine hydroxylase at 2.3 Å and its implications for inherited neurodegenerative diseases. Nat Struct Mol Biol 4, 578–585 (1997). https://doi.org/10.1038/nsb0797-578
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0797-578
This article is cited by
-
DOPAnization of tyrosine in α-synuclein by tyrosine hydroxylase leads to the formation of oligomers
Nature Communications (2022)
-
Perillyl Alcohol Mitigates Behavioural Changes and Limits Cell Death and Mitochondrial Changes in Unilateral 6-OHDA Lesion Model of Parkinson’s Disease Through Alleviation of Oxidative Stress
Neurotoxicity Research (2020)
-
Human tyrosine hydroxylase in Parkinson’s disease and in related disorders
Journal of Neural Transmission (2019)
-
Proteasome-mediated degradation of tyrosine hydroxylase triggered by its phosphorylation: a new question as to the intracellular location at which the degradation occurs
Journal of Neural Transmission (2018)
-
Noggin Along with a Self-Assembling Peptide Nanofiber Containing Long Motif of Laminin Induces Tyrosine Hydroxylase Gene Expression
Molecular Neurobiology (2017)