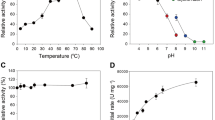
Abstract
The structure of Clostridium thermocellum endoglucanase CeIC, a member of the largest cellulase family (family A), has been determined at 2.15 Å resolution. The protein folds into an (α/β)8 barrel, with a deep active-site cleft generated by the insertion of a helical subdomain. The structure of the catalytic core of xylanase XynZ, which belongs to xylanase family F, has been determined at 1.4 Å resolution. In spite of significant differences in substrate specificity and structure (including the absence of the helical subdomain), the general polypeptide folding pattern, architecture of the active site and catalytic mechanism of XynZ and CeIC are similar, suggesting a common evolutionary origin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
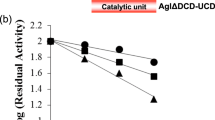
References
Gilkes, N.R., Henrissat, B., Kilburn, D.G., Miller, R.C., Jr & Warren, R.A.J. Domains in microbial β-1,4-glycanases: Sequence conservation, function, and enzyme families. Microbiol. Rev. 55, 303–315 (1991).
Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 (1991).
Henrissat, B. & Bairoch, A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293, 781–788 (1993).
Gebler, J. et al. Stereoselective hydrolysis catalyzed by related β-1,4-glucanases and β-1,4-xylanases. J. biol. Chem. 267, 12559–12561 (1992).
Banner, D.W. et al. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 Å resolution using amino acid sequence data. Nature 255, 609–614 (1975).
Wang, Q. et al. Glu 280 is the nucleophile in the active site of Clostridium thermocellum CelC, a family A endo-β-1,4-glucanase. J. biol. Chem. 268, 14096–14102 (1993).
Navas, J. & Béguin, P. Site-directed mutagenesis of conserved residues in Clostridium thermocellum endoglucanase CelC. Biochem. biophys. res. Comm. 189, 807–812 (1992).
Py, B., Bortoli-German, I., Haiech, J., Chippaux, M. & Barras, F. Cellulase EGZ of Erwinia chrysanthemi: structural organization and importance of His 98 and Glu 133 residues for catalysis. Prot. Engng 4, 325–333 (1991).
Belaich, A. et al. The catalytic domain of endoglucanase A from Clostridium cellulolyticum - Effects of arginine-79 and histidine-122 mutations on catalysis. J. Bacteriol. 174, 4677–4682 (1992).
Davies, G.J. et al. Structure and function of endoglucanase V. Nature 365, 362–364 (1993).
Törrönen, A., Harkki, A. & Rouvinen, J. Three-dimensional structure of endo-1,4-β-xylanase II from Trichoderma reesei: Two conformational states in the active site. EMBO J. 13, 2493–2501 (1994).
Derewenda, U. et al. Crystal structure, at 2.6 Å resolution, of the Streptomyces lividans xylanase A, a member of the F family of β-1,4-D-glycanases. J. biol. Chem. 269, 20811–20814 (1994).
White, A., Withers, S.G., Gilkes, N.R. & Rose, D.R. Crystal structure of the catalytic domain of the β-1,4-glycanase Cex from Cellulomonas fimi. Biochemistry 33, 12546–12552 (1994).
Harris, G.W. et al. Structure of the catalytic core of the family F xylanase from Pseudomonas fluorescens and identification of the xylopentaose-binding sites. Structure 2 1107–1116 (1994).
Tull, D., Withers, S.G., Gilkes, N.R., Kilburn, D.G., Warren, R.A.J. & Aebersold, R. Glutamic acid 274 is the nucleophile in the active site of a “retaining” exoglucanase from Cellulomonas fimi. J. biol. Chem. 266, 15621–15625 (1991).
Varghese, J.N., Garrett, T.P.J., Colman, P.M., Chen, L., HøJ, P.B. & Fincher, G.B. Three-dimensional structure of two plant beta-glucan endohydrolases with distinct substrate specificities. Proc. natn. Acad. Sci. U.S.A. 91, 2785–2789 (1994).
Chen, L., Fincher, G.B. & HøJ, P.B. Evolution of polysaccharide hydrolase substrate specificity. J. biol. Chem. 268, 13318–13326 (1993).
Jenkins, J., Lo Leggio, L., Harris, G. & Pickersgill, R. β-glucosidase, β-galactosidase, family A cellulases, family F xylanases and two barley glycanases form a superfamily of enzymes with 8-fold β/α architecture and with two conserved glutamates near the carboxy-terminal ends of β-strands four and seven. FEBS Let. in the press.
Fierobe, H.-P. et al. Characterization of endoglucanase A from Clostridium cellulolyticum. J. Bacteriol. 173 7956–7962. (1991).
Yagüe, E., Béguin, P. & Aubert, J.-P. Nucleotide sequence and deletion analysis of the cellulase-encoding gene celH of Clostridium thermocellum. Gene 89, 61–67 (1990).
Gilkes, N.R., Langsford, M.L., Kilburn, D.G., Miller, Jr, R.C. & Warren, R.A.J. Mode of action and substrate specificities of cellulases from cloned bacterial genes. J. biol. Chem. 259, 10455–10459 (1984).
Grépinet, O., Chebrou, M.-C. & Béguin, P. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J. Bacteriol. 170, 4582–4588 (1988).
Claeyssens, M. & Henrissat, B. Specificity mapping of cellulolytic enzymes - classification into families of structurally related proteins confirmed by biochemical analysis. Prot. Sci. 1, 1293–1297 (1992).
Henrissat, B., Callebaut, I., Fabrega, S., Lehn, P., Mornon, J.-P. & Davies, G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. natn. Acad. Sci. U.S.A., in the press.
Rouvinen, J., Bergfors, T., Teeri, T., Knowles, J.K.C. & Jones, T.A. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249, 380–386 (1990).
Divne, C. et al. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265, 524–528 (1994)
Juy, M. et al. Crystal structure of a thermostable bacterial cellulose-degrading enzyme. Nature 357, 89–91 (1992).
Souchon, H., Spinelli, S., Béguin, P. & Alzari, P.M. Crystallization and preliminary diffraction analysis of the catalytic domain of xylanase Z from Clostridium thermocellum. J. molec. Biol. 235, 1348–1350 (1994).
Dominguez, R., Souchon, H. & Alzari, P.M. Characterization of two crystal forms of Clostridium thermocellum endoglucanase CelC. Proteins 19, 158–160 (1994).
CCP4, The SERC (UK) Collaborative Computing Project No. 4: A Suite of Programs for Protein Crystallography (Daresbury, UK; 1979).
Navaza, J. AMoRe: an automated package for molecular replacement. Acta crystallogr. A50, 157–163 (1994).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta crystallogr. A47, 110–119 (1991).
Brünger, A.T., Kuriyan, J. & Karplus, M., R-factor refinement by molecular dynamics. Science 235, 458–460 (1987).
Engh, R.A. & Huber, R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta crystallogr. A47, 392–400 (1991).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. appl. Crystallogr. 26, 283–291 (1993).
Sheldrick, G.M. Phase annealing in SHELX-90: direct methods for larger structures. Acta crystallogr. A46, 467–473 (1990).
Lamzin, V.S. & Wilson, K.S. Automated refinement of protein models. Acta crystallogr. D49, 129–147 (1993).
Kabsch, W. A solution for the best rotation to relate two sets of vectors. Acta crystallogr. A32, 922–923 (1976).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. appl. Crystallogr. 24, 946–950 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Dominguez, R., Souchon, H., Spinelli, S. et al. A common protein fold and similar active site in two distinct families of β-glycanases. Nat Struct Mol Biol 2, 569–576 (1995). https://doi.org/10.1038/nsb0795-569
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0795-569
This article is cited by
-
Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis
Applied Microbiology and Biotechnology (2023)
-
Microbial xylanases and their industrial application in pulp and paper biobleaching: a review
3 Biotech (2017)
-
Shifting the optimum pH of Bacillus circulans xylanase towards acidic side by introducing arginine
Biotechnology and Bioprocess Engineering (2013)
-
Characterization of a Paenibacillus cell-associated xylanase with high activity on aryl-xylosides: a new subclass of family 10 xylanases
Applied Microbiology and Biotechnology (2003)
-
Structural evidence for dimerization-regulated activation of an integral membrane phospholipase
Nature (1999)