Abstract
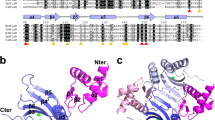
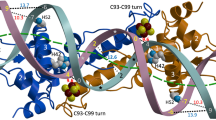
The arginine repressor (ArgR) is a hexameric DNA-binding protein that plays a multifunctional role in the bacterial cell. Here, we present the 2.5 Å structure of apo-ArgR from Bacillus stearothermophilus and the 2.2 Å structure of the hexameric ArgR oligomerization domain with bound arginine. This first view of intact ArgR reveals an approximately 32-symmetric hexamer of identical subunits, with six DNA-binding domains surrounding a central oligomeric core. The difference in quaternary organization of subunits in the arginine-bound and apo forms provides a possible explanation for poor operator binding by apo-ArgR and for high affinity binding in the presence of arginine.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Glansdorff, N. in Escherichia coli and Salmonella: cellular and molecular biology. (ed. Neidhardt, F.C.) 408–433 (American Society for Microbiology Press, Washington, D.C.; 1996).
Maas, W.K. Microbiol. Rev. 58, 631–640 (1994).
Czaplewski, L.G., North, A.K., Smith, M.C.M., Baumberg, S. & Stockley, P.G. Mol. Microbiol. 6 , 267–275 (1992).
Stirling, C.J., Szatmari, G., Stewart, G., Smith, M.C. & Sherratt, D.J. EMBO J. 7, 4389–4395 (1988).
Lim, D.B., Oppenheim, J.D., Eckhardt, T. & Maas, W.K. Proc. Natl. Acad. Sci. USA 84, 6697– 701 (1987).
North, A.K., Smith, M.C. & Baumberg, S. Gene 80, 29– 38 (1989).
Lu, C.D., Houghton, J.E. & Abdelal, A.T. J. Mol. Biol. 225, 11– 24 (1992).
Dion, M. et al. Mol. Microbiol. 25, 385– 398 (1997).
Fleischmann, R.D. et al. Science 269, 496–512 (1995).
Cole, S.T. et al. Nature 393, 537–544 (1998).
Rodríguez–García, A., Ludovice, M., Martín, J.F. & Liras, P. Mol. Microbiol. 25, 219– 228 (1997).
Tian, G. & Maas, W.K. Mol. Microbiol. 13, 599–608 (1994).
Burke, M., Merican, A.F. & Sherratt, D.J. Mol. Microbiol. 13, 609– 618 (1994).
Van Duyne, G.D., Ghosh, S., Maas, W.K. & Sigler, P.B. J. Mol. Biol. 256, 377–391 ( 1996).
Grandori, R. et al. J. Mol. Biol. 254, 150– 162 (1995).
Chen, S.H., Merican, A.F. & Sherratt, D.J. Mol. Microbiol. 24, 1143– 1156 (1997).
Sunnerhagen, M., Nilges, M., Otting, G. & Carey, J. Nature Struct. Biol. 4, 819–826 ( 1997).
Brennen, R.G. Cell 74, 773–776 ( 1993).
Tian, G., Lim, D., Oppenheim, J.D. & Maas, W.K. J. Mol. Biol. 235, 221–230 (1994).
Wang, H., Glansdorff, N. & Charlier, D. J. Mol. Biol. 277, 805– 824 (1998).
Tian, G., Lim, D., Carey, J. & Maas, W.K. J. Mol. Biol. 226, 387–397 (1992).
Charlier, D. et al. J. Mol. Biol. 226, 367– 386 (1992).
Schultz, S.C., Shields, G.C. & Steitz, T.A. Science 253, 1001– 1007 (1991).
Miller, C.M., Baumberg, S. & Stockley, P.G. Mol. Microbiol. 26, 37– 48 (1997).
Abrahams, J.P. & Leslie, A.G.W. Acta Crystallogr. D 52, 30–42 ( 1996).
Navaza, J. Acta Crystallgr. A 50, 157–163 (1994).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Acta Crystallogr. A 47, 110–119 ( 1991).
Otwinowski, Z. in CCP4 Proceedings. 80–88 (Daresbury Laboratory, Warrington, UK, 1991).
Jiang, J.S. & Brunger, A.T. J. Mol. Biol. 243, 100–115 (1994).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. J. Appl. Crystallogr. 26, 283–291 (1993).
Kraulis, P. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E.A. & Bacon, D.J. Methods Enzymol. 277 , 505–524 (1997).
Carson, M. J. Appl. Crystallgr. 24, 958–961 (1991).
Acknowledgements
We thank the staff of the CHESS F1 beamline for synchrotron support, F. Guo and M. Gopaul for assistance with data collection, and X. Li for help in crystallization. We also acknowledge helpful comments and discussions from M. Lemmon, H. Lu, H. Nelson, M. Lewis, and W.K. Maas. Supported by a grant from the National Institutes of Health to G.V.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ni, J., Sakanyan, V., Charlier, D. et al. Structure of the arginine repressor from Bacillus stearothermophilus . Nat Struct Mol Biol 6, 427–432 (1999). https://doi.org/10.1038/8229
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/8229
This article is cited by
-
Regulation of arginine biosynthesis, catabolism and transport in Escherichia coli
Amino Acids (2019)
-
Binding-competent states for L-arginine in E. coli arginine repressor apoprotein
Journal of Molecular Modeling (2014)
-
Genome-wide comprehensive analysis of transcriptional regulation by ArgR in Thermus thermophilus
Extremophiles (2014)
-
Structural basis of lipid biosynthesis regulation in Gram-positive bacteria
The EMBO Journal (2006)
-
Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains
Applied Microbiology and Biotechnology (2006)