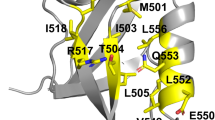
Abstract
The prevailing view in the field of protein folding holds that the native state is the most stable structure possible. A corollary of this thermodynamic hypothesis is that the native state is in equilibrium with all other conformations of the protein. We have found an example of a protein that may exist in two different states, both of which may be regarded as ‘native’, but which cannot equilibrate on a timescale that is biologically meaningful. We propose that the active conformation of this protein is at only one of several energy minima, and that during the process of refolding in vitro — and, we assume, folding in vivo — the choice of which state the polypeptide finally attains is determined by kinetic partitioning between folding pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Lau, K.F. & Dill, K.A. Theory for protein mutability and biogenesis. Proc. natn. Acad. Sci. U.S.A. 87, 638–642 (1990).
Matthews, C.R. Pathways of protein folding. A. Rev. Biochem. 62, 653–683 (1993).
Waddle, J.J., Johnston, T.C. & Baldwin, T.O. Polypeptide folding and dimerization in bacterial luciferase occur by a concerted mechanism in vivo. Biochemistry 26, 4917–4921 (1987).
Sugihara, J. & Baldwin, T.O. Effects of 3′ end deletions from the Vibrio harveyi luxB gene on luciferase subunit folding and enzyme assembly: generation of temperature-sensitive polypeptide folding mutants. Biochemistry 27, 2872–2880 (1988).
Ziegler, M.M., Goldberg, M.E., Chaffotte, A.-F. & Baldwin, T.O. Refolding of luciferase subunits from urea and assembly of the active heterodimer: evidence for folding intermediates that precede and follow the dimerization step on the pathway to the active form of the enzyme. J. biol. Chem. 268, 10760–10765 (1993).
Baldwin, T.O. Ziegler, M.M., Chaffotte, A.-F. & Goldberg, M.E. Contribution of folding steps involving the individual subunits of bacterial luciferase to the assembly of the active heterodimeric enzyme. J. biol. Chem. 268, 10766–10772 (1993).
Clark, A.C., Sinclair, J.F. & Baldwin, T.O. Folding of bacterial luciferase involves a non-native heterodimeric intermediate in equilibrium with the native enzyme and the unfolded subunits. J. biol. Chem. 268, 10773–10779 (1993).
Sinclair, J.F., Waddle, J.J., Waddill, E.F. & Baldwin, T.O. Purified native subunits of bacterial luciferase are active in the bioluminescence reaction but fail to assemble into the αβ structure. Biochemistry 32, 5036–5044 (1993).
Wetlaufer, D.B. Folding of protein fragments. Adv. Protein Chem. 34, 61–92 (1981).
Waddle, J. & Baldwin, T.O. Individual α and β subunits of bacterial luciferase exhibit bioluminescence activity. Biochem. biophys. Res. Commun. 178, 1188–1193 (1991).
Johnston, T.C., Thompson, R.B. & Baldwin, T.O. Nucleotide sequence of the luxB gene of Vibrio harveyi and the complete amino acid sequence of the β subunit of bacterial luciferase. J. biol. Chem. 261, 4805–4811 (1986).
Waddle, J.J. Purification of bacterial luciferase native subunits and investigation of their folding, association and activity (Ph.D. thesis, Texas A&M University, College Station, Texas, 1990).
Goldberg, M.E. The second translation of the genetic message: protein folding and assembly. Trends biochem. Sci. 10, 388–391 (1985).
Fane, B., Villafane, R., Mitraki, A. & King, J. Identification of global suppressors for temperature sensitive folding mutations of the P22 tailspike protein. J. biol. Chem. 266, 11640–11648 (1991).
Goldenberg, D.P., Smith, D.H. & King, J. Genetic analysis of the folding pathway for the tail spike protein of phage P22. Proc. natn. Acad. Sci. U.S.A. 80, 7060–7064 (1983).
Baker, D., Sohl, J.L. & Agard, D.A. A protein-folding reaction under kinetic control. Nature 356, 263–265 (1992).
Mottonen, J., Strand, A., Symersky, J., Sweet, R.M., Danley, D.E., Geoghegan, K.F., Gerard, R.D. & Goldsmith, E.J. Structural basis of latency in plasminogen activator inhibitor-1. Nature 355, 270–273 (1992).
Gething, M.-J., McCammon, K. & Sambrook, J. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell 46, 939–950 (1986).
Eder, J., Rheinnecker, M. & Fersht, A.R. Folding of subtilisin BPN': characterization of a folding intermediate Biochemistry 32, 18–26 (1993).
Hastings, J.W., Baldwin, T.O. & Nicoli, M.Z. Bacterial luciferase: assay, purification, and properties. Meth. Enzymol. 57, 135–152 (1978).
Baldwin, T.O., Chen, L.H., Chlumsky, L.J., Devine, J.H. & Ziegler, M.M. Site-directed mutagenesis of bacterial luciferase: analysis of the ‘essential’ thiol. J. biol. Chem. 4, 40–48 (1989).
Tu, S.-C. Preparation of the subunits of bacterial luciferase. Meth. Enzymol. 62, 171–174(1978).
Baldwin, T.O. & Ziegler, M.M. in Chemistry and Biochemistry of Flavoenzymes (ed. Müller, F.) Vol. III, 467–530 (CRC Press, Boca Raton, Florida, 1992).
Kratky, O., Leopold, H & Stabinger, H. The determination of the partial specific volume of proteins by the mechanical oscillator technique. Meth. Enzym. 27, 98–110 (1973).
Cohn, E.J. & Edsall, J.T. in Proteins, Amino Acids and Peptides (Van Nostrand-Reinhold, Princeton, New Jersey, 1943).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Sinclair, J., Ziegler, M. & Baldwin, T. Kinetic partitioning during protein folding yields multiple native states. Nat Struct Mol Biol 1, 320–326 (1994). https://doi.org/10.1038/nsb0594-320
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0594-320
This article is cited by
-
Thermodynamic and kinetic stability of a large multi-domain enzyme from the hyperthermophile Aeropyrum pernix
Extremophiles (2010)
-
Physiological responses of Escherichia coli exposed to different heat-stress kinetics
Archives of Microbiology (2010)