Abstract
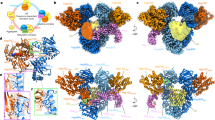
Heat shock protein 33 (Hsp33) inhibits aggregation of partially denatured proteins during oxidative stress. The chaperone activity of Hsp33 is unique among heat shock proteins because the activity is reversibly regulated by cellular redox status. We report here the crystal structure of the N-terminal region of Hsp33 fragments with constitutive chaperone activity. The structure reveals that the N-terminal portion of Hsp33 forms a tightly associated dimer formed by a domain crossover. A concave groove on the dimeric surface contains an elongated hydrophobic patch that could potentially bind denatured protein substrates. The termini of the subunits are located near the hydrophobic patch, indicating that the cleaved C-terminal domain may shield the hydrophobic patch in an inactive state. Two of the four conserved zinc-coordinating cysteines are in the end of the N-terminal domain, and the other two are in the cleaved C-terminal domain. The structural information and subsequent biochemical characterizations suggest that the redox switch of Hsp33 occurrs by a reversible dissociation of the C-terminal regulatory domain through oxidation of zinc-coordinating cysteines and zinc release.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Halliwell, B. & Gutteridge, J.M.C. Free radicals in biology and medicine, third edition (Oxford University Press, New York; 1999).
Lander, H.M. An essential role for free radicals and derived species in signal transduction. FASEB J. 11, 118–124 (1997).
Adler, V., Yin, Z., Tew, K.D. & Ronai, Z. Role of redox potential and reactive oxygen species in stress signaling. Oncogene 18, 6104–6111 (1999).
Jakob, U., Muse, W., Eser, M. & Bardwell, J.C.A. Chaperone activity with a redox switch. Cell 96, 341–352 (1999).
Ruddock, L.W. & Klappa, P. Oxidative stress: protein folding with a novel redox switch. Curr. Biol. 3, R400–R402 (1999).
Barbirz, S. Jakob, U. & Glocker, M.O. Mass spectrometry unravels disulfide bond formation as the mechanism that activates a molecular chaperone. J. Biol. Chem. 275, 18759–18766 (2000).
Hendrickson, W.A. & Ogata, C.M. Phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol. 276, 494–523 (1997).
Ormö, M. et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273, 1392–1395 (1996).
Boggon, T.J., Shan, W.-S., Santagata, S., Myers, S.C. & Shapiro, L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286, 2119–2125 (1999).
Xu, Z., Horwich, A.L. & Sigler, P.B. The crystal structure of the asymmetric GroEL–GroES–(ADP)7 chaperone complex. Nature 388, 741–750 (1997).
Kim, K.K., Kim, R. & Kim, S.-H. Crystal structure of a small heat-shock protein. Nature 394, 595–599 (1998).
Zhu, X. et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 (1996).
Leroux, M.R., Melki, R., Gordon, B., Batelier, G. & Candido, E.P. Structure-function studies on small heat shock protein oligomeric assembly and interaction with unfolded polypeptides. J. Biol. Chem. 272, 24646–24656 (1997).
Chadli, A., Ladjimi, M.M., Baulieu, E.E. & Catelli, M.G. Heat-induced oligomerization of the molecular chaperone Hsp90. Inhibition by ATP and geldanamycin and activation by transition metal oxyanions. J. Biol. Chem. 274, 4133–4139 (1999).
Schlunegger, M.P., Bennett, M.J. & Eisenberg, D. Oligomer formation by 3D domain swapping: a model for protein assembly and misassembly. Adv. Protein Chem. 50, 61–122 (1997).
Bennett, M.J. & Eisenberg, D. Refined structure of monomeric diphtheria toxin at 2.3 Å resolution. Protein Sci. 3, 1464–1475 (1994).
Norledge, B.V. et al. The X-ray structures of two mutant crystallin domains shed light on the evolution of multi-domain proteins. Nature Struct. Biol. 3, 267–274 (1996).
Milburn, M.V. et al. A novel dimer configuration revealed by the crystal structure at 2.4 Å resolution of human interleukin-5. Nature 363, 172–176 (1993).
Jakob, U., Eser, M. & Bardwell, J.C.A. Hsp33's redox switch has a novel zinc-binding motif. J. Biol. Chem. 275, 38302–38310 (2000).
Aslund, F. & Beckwith, J. Bridge over troubled waters: sensing stress by disulfide bond formation. Cell 96, 751–753 (1999).
Zheng, M., Aslund, F. & Storz, G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721 (1998).
Saitoh, M. et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17, 2596–2606 (1998).
Toledano, M.B. et al. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78, 897–909 (1994).
Gotoh, Y. & Cooper, J.A. Reactive oxygen species- and dimerization-induced activation of apoptosis signal-regulating kinase 1 in tumor necrosis factor-alpha signal transduction. J. Biol. Chem. 273, 17477–17482 (1998).
Choi, H.-J. et al. Structural basis of the redox switch in the OxyR transcription factor. Cell in the press (2001).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Terwilliger, T.C. & Berendzen, J. Automated structure solution for MIR and MAD. Acta Crystallogr. D 55, 849–861 (1999).
Brünger, A.T. et al. Crystallography & NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Carson, M. Ribbons. Methods Enzymol. 277, 493–505 (1997).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Esnouf, R.M. An extensively modified version of MolScript that includes greatly enhanced colouring capabilities. J. Mol. Graph. Model. 15, 133–138 (1997).
Hensley, P. Defining the structure and stability of macromolecular assemblies in solution: the re-emergence of analytical ultracentrifugation as a practical tool. Structure 4, 367–373 (1996).
Gill, S.C. & von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 (1989).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid. Res. 22, 4673–4680 (1994).
Barton, G. J. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 6, 37–40 (1993).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Acknowledgements
We gratefully acknowledge C. Ogata and the staffs in the NSLS beamline X4A for help with data collection. This work was supported by a national creative research initiatives grant from the ministry of science and technology, Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, SJ., Jeong, DG., Chi, SW. et al. Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity. Nat Struct Mol Biol 8, 459–466 (2001). https://doi.org/10.1038/87639
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/87639
This article is cited by
-
The redox-switch domain of Hsp33 functions as dual stress sensor
Nature Structural & Molecular Biology (2007)