Abstract
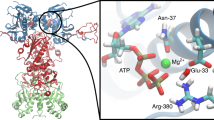
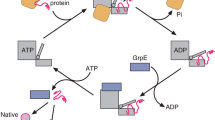
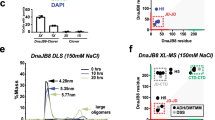
The Hsp70 chaperone activity in protein folding is regulated by ATP-controlled cycles of substrate binding and release. Nucleotide exchange plays a key role in these cycles by triggering substrate release. Structural searches of Hsp70 homologs revealed three structural elements within the ATPase domain: two salt bridges and an exposed loop. Mutational analysis showed that these elements control the dissociation of nucleotides, the interaction with exchange factors and chaperone activity. Sequence variations in the three elements classify the Hsp70 family members into three subfamilies, DnaK proteins, HscA proteins and Hsc70 proteins. These subfamilies show strong differences in nucleotide dissociation and interaction with the exchange factors GrpE and Bag-1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hartl, F.U. Nature 381, 571–580 (1996).
Bukau, B. & Horwich, A.L. Cell 92, 351–366 (1998).
Schmid, D., Baici, A., Gehring, H. & Christen, P. Science 263, 971–973 (1994).
Mayer, M.P. et al. Nature Struct. Biol. 7, 586–593 (2000).
Theyssen, H., Schuster, H.-P., Bukau, B. & Reinstein, J. J. Mol. Biol. 263, 657–670 (1996).
Ha, J.-H. & McKay, D.B. Biochemistry 33, 14625–14635 (1994).
Ha, J.-H. & McKay, D.B. Biochemistry 34, 11635–11644 (1995).
Slepenkov, S.V. & Witt, S.N. Biochemistry 37, 1015–1024 (1998).
Russell, R., Jordan, R. & McMacken, R. Biochemistry 37, 596–607 (1998).
Liberek, K., Marszalek, J., Ang, D., Georgopoulos, C. & Zylicz, M. Proc. Natl. Acad. Sci. USA 88, 2874–2878 (1991).
Packschies, L. et al. Biochemistry 36, 3417–3422 (1997).
Bimston, D. et al. EMBO J. 17, 6871–6878 (1998).
Takayama, S. et al. EMBO J. 16, 4887–4896 (1997).
Höhfeld, J. & Jentsch, S. EMBO J. 16, 6209–6216 (1997).
Silberg, J.J. & Vickery, L.E. J. Biol. Chem. 275, 7779–7786 (2000).
Harrison, C.J., Hayer-Hartl, M., Di Liberto, M., Hartl, F.-U. & Kuriyan, J. Science 276, 431–435 (1997).
Flaherty, K.M., Deluca-Flaherty, C. & McKay, D.B. Nature 346, 623–628 (1990).
Buchberger, A. et al. J. Biol. Chem. 270, 16903–16910 (1995).
McCarty, J.S. et al. J. Mol. Biol. 256, 829–837 (1996).
Schröder, H., Langer, T., Hartl, F.-U. & Bukau, B. EMBO J. 12, 4137–4144 (1993).
Szabo, A. et al. Proc. Natl. Acad. Sci. USA 91, 10345–10349 (1994).
Kunkel, T.A., Bebenek, K. & McClary, J. Methods. Enzymol. 204, 125–139 (1991).
Bukau, B. & Walker, G. EMBO J. 9, 4027–4036 (1990).
Buchberger, A., Schröder, H., Büttner, M., Valencia, A. & Bukau, B. Nature Struct. Biol. 1, 95–101 (1994).
Hesterkamp, T. & Bukau, B. EMBO J. 17, 4818–4828 (1998).
Schönfeld, H.-J., Schmidt, D. & Zulauf, M. Progr. Colloid. Polym. Sci. 99, 7–10 (1995).
Peitsch, M.C. Bio/Technology 13, 658–660 (1995).
Peitsch, M.C. Biochem. Soc. Trans. 24, 274–279 (1996).
Guex, N. & Peitsch, M.C. Electrophoresis 18, 2714–2723 (1997).
McCarty, J.S., Buchberger, A., Reinstein, J. & Bukau, B. J. Mol. Biol. 249, 126–137 (1995).
Acknowledgements
We thank J. Höhfeld and R. Morimoto for plasmids expressing Bag-1M and Hsc70; A. Valencia and A. Buchberger for design and cloning of dnaK-K55A; T. Hesterkamp, A. Hoelz and T. Laufen for helpful discussions. This work was supported by grants of the DFG to J.R., and the DFG (Graduiertenkolleg Biochemie der Enzyme; Leibniz program) and the Fonds der Chemischen Industrie to B.B.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brehmer, D., Rüdiger, S., Gässler, C. et al. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Mol Biol 8, 427–432 (2001). https://doi.org/10.1038/87588
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/87588
This article is cited by
-
Disease-associated mutations within the yeast DNAJB6 homolog Sis1 slow conformer-specific substrate processing and can be corrected by the modulation of nucleotide exchange factors
Nature Communications (2022)
-
Molecular characterization and transcriptional modulation of stress-responsive genes under heavy metal stress in freshwater ciliate, Euplotes aediculatus
Ecotoxicology (2022)
-
Hsp90 co-chaperones, FKBP52 and Aha1, promote tau pathogenesis in aged wild-type mice
Acta Neuropathologica Communications (2021)
-
Targeting Hsp70 facilitated protein quality control for treatment of polyglutamine diseases
Cellular and Molecular Life Sciences (2020)
-
A novel variant of the human mitochondrial DnaJ protein, Tid1, associates with a human disease exhibiting developmental delay and polyneuropathy
European Journal of Human Genetics (2019)