Abstract
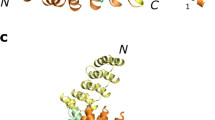
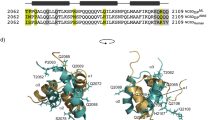
The DNA-binding domain of c-Myb consists of three imperfect tandem repeats (R1, R2 and R3). The three repeats have similar overall architectures, each containing a helix-turn-helix variation motif. The three conserved tryptophans in each repeat participate in forming a hydrophobic core. Comparison of the three repeat structures indicated that cavities are found in the hydrophobic core of R2, which is thermally unstable. On complexation with DNA, the orientations of R2 and R3 are fixed by tight binding and their conformations are slightly changed. No significant changes occur in the chemical shifts of R1 consistent with its loose interaction with DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Graf T. Myb: A transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr. Opin. Gen. Dev. 2, 249–255 (1992).
Lüscher, B. & Eisenman, R.N. New light on Myc and Myb. PartII. Myb. Genes Dev. 4, 2235–2241 (1990).
Biedenkapp, H., Borgmeyer, U., Sippel, A.E. & Klempnauer, K.-H. Viral myb oncogene encodes a sequence-specific DNA binding activity. Nature 335, 835–837 (1988).
Weston, K. Extension of the DNA binding consensus of the chicken c-Myb and v-Myb proteins. Nucleic Acids Res. 20, 3043–3049 (1992).
Tanikawa, J. et al. Recognition of specific DNA sequences by the c-myb proto-oncogene product-role of three repeat units in the DNA-binding domain. Proc. natn. Acad. Sci. U.S.A. 90, 9320–9324 (1993).
Sakura, H. et al. Delineation of three functional domains of the transcriptional activator encoded by the c-myb proto-oncogene. Proc. natn. Acad. Sci. U.S.A. 86, 5758–5762 (1989).
Gonda, T.J., Gough, N.M., Dunn, A.R. & de Blaquiere, J. Nucleotide sequence of cDNA clones of the murine myb proto-oncogene. EMBO J. 4, 2003–2008 (1985).
Klempnauer, K.-H. & Sippel, A.E. The highly conserved amino-terminal region of the protein encoded by the v-myb oncogene functions as a DNA-binding domain. EMBO J. 6, 2719–2725 (1987).
Anton, I.A. & Frampton, J. Tryptophans in myb proteins. Nature 336, 719 (1988).
Frampton, J., Gibson, T.J., Ness, S.A., Doderlein, G. & Graf, T. Proposed structure for the DNA-binding domain of the Myb oncoprotein based on model building and mutational analysis. Prot. Engng 4, 891–901 (1991).
Kanei-Ishii, C. et al. The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J. Biol. Chem. 265, 19990–19995 (1990).
Saikumar, P., Murali, R. & Reddy, E.P. Role of tryptophan repeats and flanking amino acids in Myb-DNA interactions. Proc. natn. Acad. Sci. U.S.A. 87, 8452–8456 (1990).
Gabrielsen, O.S., Sentenac, A. & Fromageot, P. Specific DNA binding by c-Myb: Evidence for a double helix-turn-helix-related motif. Science 253, 1140–1143 (1991).
Sarai, A. et al. Thermal stability of the DNA-binding domain of the Myb oncoprotein. Biochemistry 32, 7759–7764 (1993).
Jamin, N., Gabrielsen, O.S., Gilles, N., Lirsac, P.-N. & Toma, F. Secondary structure of the DNA-binding domain of the c-Myb oncoprotein in solution. Eur. J. Biochem. 216, 147–154 (1993).
Myrset, A.M. et al. DNA and redox state induced conformational changes in the DNA-binding domain of the myb oncoprotein. EMBO J. 12, 4625–4633 (1993).
Patel, L., Abate, C. & Curran, T. Altered protein conformation on DNA binding by Fos and Jun. Nature 347, 572–574 (1990).
Ogata, K. et al. Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. natn. Acad. Sci. U.S.A. 89, 6428–6432 (1992).
Harrison, S.C. & Aggarwal, A.K. DNA recognition by proteins with the helix-turn-helix motif. A. Rev. Biochem. 59, 933–969 (1990).
Pabo, C.O. & Sauer, R.T. Transcription factors: Structural families and principles of DNA recognition. A. Rev. Biochem. 61, 1053–1095 (1992).
Ogata, K. et al. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79, 639–648 (1994).
Hojo, H. & Aimoto, S. Polypeptide synthesis using the S-alkyl thioester of a partially protected peptide segment. Synthesis of the DNA-binding domain of c-Myb protein (142-193)-NH2 . Bull. chem. Soc. Jpn 64, 111–117 (1991).
Bender, T.P. & Kuehl, W.M. Murine myb protooncogene mRNA: cDNA sequence and evidence for 5′ heterogeneity. Proc. natn. Acad. Sci. U.S.A. 83, 3204–3208 (1986).
Nakai, T., Kidera, A. & Nakamura, H. Intrinsic nature of the three-dimensional structure of proteins as determined by distance geometry with good sampling properties. J. biomolec. NMR 3, 19–40 (1993).
Brennan, R.G. & Matthews, B.W. The helix-turn-helix DNA binding motif. J. biol. Chem. 264, 1903–1906 (1989).
Brennan, R.G. The winged-helix DNA-binding motif: Another helix-turn-helix takeoff. Cell 74, 773–776 (1993).
Introna, M., Golay, J., Frampton, J., Nakano, T., Ness, S.A. & Graf, T. Mutations in v-myb alter the differentiation of myelomonocytic cells transformed by the oncogene. Cell 63, 1287–1297 (1990).
Ness, S.A., Marknell, A. & Graf, T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 59, 1115–1125 (1989).
Ness, S.A., Kowentz-Leutz, E., Casini, T., Graf, T. & Leutz, A. Myb and NF-M: combinatorial activators of myeloid genes in heterologous cell types. Gene Dev. 7, 749–759 (1993).
Burk, O., Mink, S., Ringwald, M., and Klempnauer, K.-H. Synergistic activation of the chicken mim-1 gene by v-myb and C/EBP transcription factors. EMBO J. 12, 2027–2038 (1993).
Pavletich, N.P. & Pabo, C.O. Crystal structure of a five-finger GLI-DNA complex: New perspectives on zinc fingers. Science 261, 1701–1707 (1993).
Klempnauer, K.-H., Gonda, T.J. & Bishop, M.J. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb the architecture of a transduced oncogene. Cell 31, 453–463 (1982).
Dini, P.W. & Lipsick, J.S. Oncogenic truncation of the first repeat of c-Myb decreases DNA binding in vitro and in vivo. Molec. cell. Biol. 13, 7334–7348 (1993).
Messerle, B.A., Wider, G., Otting, G., Weber, C. & Wüthrich, K. Solvent suppression using a spin lock in 2D and 3D NMR spectroscopy with H2O solutions. J. magn. Reson. 85, 608–613 (1989).
Kay, L.E., Marion, D. & Bax, A. Practical aspects of 3D heteronuclear NMR of proteins. J. magn. Reson. 84, 72–84 (1989).
Marion, D. et al. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence spectroscopy: Application to interleukin 1β. Biochemistry 28, 6150–6156 (1989).
Frenkiel, T., Bauer, C., Carr, M.D., Birdsall, B. & Feeney, J. HMQC-NOESY-HMQC, a three-dimensional NMR experiment which allows detection of nuclear Overhauser effects between protons with overlapping signals. J. magn. Reson. 90, 420–425 (1990).
Ikura, M., Bax, A., Clore, G.M. & Gronenborn, A.M. Detection of nuclear Overhauser effects between degenerate amide proton resonances by heteronuclear three-dimensional nuclear magnetic resonance spectroscopy. J. Am. chem. Soc. 112, 9020–9022 (1990).
Rance, M. et al. Improved spectral resolution in COSY 1H NMR spectra of proteins via double quantum filtering. Biochem. biophys. Res. Commun. 117, 479–485 (1983).
Macura, S. & Ernst, R.R. Elucidation of cross relaxation in liquids by two-dimensional NMR spectroscopy. Molec. Phys. 41, 95–117 (1980).
Bax, A. & Davis, D.G. MLEV-17 based two dimensional homonuclear magnetization transfer spectroscopy. J. magn. Reson. 65, 355–360 (1985).
Griesinger, C., Otting, G., Wüthrich, K. & Ernst, R.R. Clean TOCYSY for 1H spin system identification in macromolecules. J. Am. chem. Soc. 110, 7870–7872 (1988).
Shaka, A.J., Lee, C.J. & Pines, A. Iterative schemes for bilinear operators; Application to spin decoupling. J. magn. Reson. 77, 274–293 (1988).
Cavanagh, J. & Rance, M. Suppression of cross-relaxation effects in TOCSY spectra via a modified DIPSI-2 mixing sequence. J. magn. Reson. 96, 670–678 (1992).
Rance, M. Improved techniques for homonuclear rotating-frame and isotropic mixing experiments. J. magn. Reson. 74, 557–564 (1987).
Kim, Y. & Prestegard, J.H. Measurement of vicinal couplings from cross peaks in COSY spectra. J. magn. Reson. 84, 9–13 (1989).
Driscoll, P.C., Gronenborn, A.M., Beress, L., and Clore, G.M. Determination of the three-dimensional solution structure of the antihypertensive and antiviral protein BDS-1 from the sea anemone Anemonia sulcata: A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry 28, 2188–2198 (1989).
Wagner, G. et al. Protein structures in solution by nuclear magnetic resonance and distance geometry. The polypeptide fold of the basic pancreatic trypsin inhibitor determined using two different algorithms, DISGEO and DISMAN. J. molec. Biol. 196, 611–639 (1987).
Driscoll, P.C., Clore, G.M., Beress, L. & Gronenborn, A.M. A proton nuclear magnetic resonance study of the antihypertensive and antiviral protein BDS-1 from the sea anemone Anemonia sulcata: Sequential and stereospecific resonance assignment and secondary structure. Biochemistry 28, 2178–2187 (1989).
Kochoyan, M. et al Altering zinc fingers in the human male associated protein ZFY:2D NMR structure of an even finger and implications for “Jumping-Linker” DNA recognition. Biochemistry 30, 3371–3386 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ogata, K., Morikawa, S., Nakamura, H. et al. Comparison of the free and DNA-complexed forms of the DMA-binding domain from c-Myb. Nat Struct Mol Biol 2, 309–320 (1995). https://doi.org/10.1038/nsb0495-309
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0495-309
This article is cited by
-
DlMYB113 mutation affects anthocyanin accumulation in red pericarp longan (Dimocarpus longan Lour.)
Horticulture Advances (2023)
-
Dynamic structures of intrinsically disordered proteins related to the general transcription factor TFIIH, nucleosomes, and histone chaperones
Biophysical Reviews (2022)
-
Disruption of transcription factor RhMYB123 causes the transformation of stamen to malformed petal in rose (Rosa hybrida)
Plant Cell Reports (2022)
-
The Arabidopsis thaliana transcription factor MYB59 regulates calcium signalling during plant growth and stress response
Plant Molecular Biology (2019)
-
Two genetic changes in cis-regulatory elements caused evolution of petal spot position in Clarkia
Nature Plants (2018)