Abstract
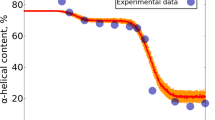
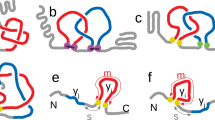
Molten globules are thought to be general intermediates in protein folding. Apparently conflicting studies have failed to clarify whether one of the best characterized molten globules, that of α-lactalbumin, resembles an expanded native-like protein or a nonspecific collapsed polypeptide. Here we show that the molten globule properties of α-lactalbumin are largely confined to one of its two domains. The α-helical domain forms a helical structure with a native-like tertiary fold, while the β-sheet domain is largely unstructured. Molten globules thus possess a native-like backbone topology, but this topology does not necessarily encompass the entire polypeptide chain. Our studies indicate that molten globules provide an approximate solution to, and considerable simplification of the protein folding problem.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Ptitsyn, O.B. The molten globule state, in Protein Folding (Ed. Creighton, T.E.) 243–300. (W.H. Freeman and Co., New York, 1993).
Bychkova, V.E. & Ptitsyn, O.B. The molten globule in vitro and in vivo. Chemtracts — Biochem. molec. Biol. 4, 133–163 (1993).
Kuwajima, K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins 6, 87–103 (1989).
Haynie, D.T. & Friere, E. Structural energetics of the molten globule state. Proteins 16, 115–140 (1993).
Christensen, H. & Pain, R.H. Molten globule intermediates and protein folding. Eur. biophys. J. 19, 221–229 (1991).
Dobson, C.M. Unfolded proteins, compact states, and molten globules. Curr. Opin. struct. Biol. 2, 6–12 (1992).
Kuwajima, K., Nitta, K., Yoneyama, M. & Sugai, S. Three-state denaturation of α-Lactalbumin by guanidine hydrochloride. J. molec. Biol. 106, 359–373 (1976).
Dolgikh, D.A. et al. α-Lactalbumin: compact state with fluctuating tertiary structure. FEBS Letts 136, 311–315 (1981).
Dolgikh, D.A. et al. Compact state of a protein molecule with pronounced small-scale mobility: bovine α-Lactalbumin. Eur. biophys. J. 13, 109–121 (1985).
Baum, J., Dobson, C.M., Evans, P.A. & Hanley, C. Characterization of a partly folded protein by NMR methods: studies on the molten globule state of guinea pig α-Lactalbumin. Biochemistry 28, 7–13 (1989).
Alexandrescu, A.T., Evans, P.A., Pitkeathly, M., Baum, J . & Dobson, C.M. Structure and dynamics of the acid-denatured molten globule state of α-Lactalbumin: a two-dimensional NMR study. Biochemistry 32, 1707–1718 (1993).
Xie, D., Bhakuni, V. & Freire, E. Calorimetric determination of the energetics of the molten globule intermediate in protein folding: Apo-α-Lactalbumin. Biochemistry 30, 10673–10678 (1991).
Ewbank, J.J. & Creighton, T.E. The molten globule protein conformation probed by disulphide bonds. Nature 350, 518–520 (1991).
Creighton, T.E. & Ewbank, J.J. Disulphide-rearranged molten globule state of α-Lactalbumin. Biochemistry 33, 1534–1538 (1994).
Peng, Z.-y. & Kim, P.S. A protein dissection study of a molten globule. Biochemistry 33, 2136–2141 (1994).
Peng, Z.-y., Wu, L.C. & Kim, P.S. Local structural preferences in the α-Lactalbumin molten globule. Biochemistry, in the press.
Redfield, C., Smith, R.A.G. & Dobson, C.M. Structural characterization of a highly-ordered ‘molten globule’ at low pH. Nature struct. Biol. 1, 23–29 (1994).
Feng, Y., Sligar, S.G. & Wand, A.J. Solution structure of apocytochrome b562. Nature struct. Biol. 1, 30–35 (1994).
Miranker, A., Robinson, C.V., Radford, S.E., Alpin, R.T. & Dobson, C.M. Detection of transient protein folding populations by mass spectrometry. Science 262, 896–900 (1993).
Radford, S.E., Dobson, C.M. & Evans, P.A. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature 358, 302–307 (1992).
Kuwajima, K., Hiraoka, Y., Ikeguchi, M. & Sugai, S. Comparison of the transient folding intermediates in lysozyme and α-Lactalbumin. Biochemistry 24, 874–881 (1985).
Ikeguchi, M., Kuwajima, K., Mitani, M. & Sugai, S. Evidence for identity between the equilibrium unfolding intermediate and a transient folding intermediate: a comparitive study of the folding reactions of α-Lactalbumin and lysozyme. Biochemistry 25, 6965–6972 (1986).
Gilmanshin, R.I. & Ptitsyn, O.B. An early intermediate of refolding α-Lactalbumin forms within 2 ms. FEBS Letters 223, 327–329 (1987).
Wetlaufer, D.B., ion, rapid folding, and globular intrachain regions in proteins. Proc. natn. Acad. Sci. U.S.A. 70, 697–701 (1973).
Jaenicke, R. Protein folding: local structures, domain, subunits, and assemblies. Biochemistry 30, 3147–3161 (1991).
Levinthal, C. Are there pathways for protein folding? J. chim. Phys. 65, 44–45 (1968).
Kunkel, T.A., Roberts, J.D. & Zakour, R.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Meths Enz. 154, 367–382 (1987).
Doering, D.S. Functional and structural studies of a small f-actin binding protein, (Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, 1992).
Studier, F.W., Rosenberg, A.H., Dunn, J.J. & Dubendorff, J.W. Use of T7 RNA polymerase to direct expression of cloned genes. Meths Enz. 185, 60–89 (1990).
Ellman, G.L. Tissue sulphydryl groups. Arch. Biochem. Biophys. 82, 70–77 (1959).
Edelhoch, H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948–1954 (1967).
Chen, Y.-H., Yang, J.T. and Chau, K.H. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry 13, 3350–3359 (1974).
Laue, T.M., Shah, B.D., Ridgeway, T.M. & Pelletier, S.L. Computer-aided interpretation of analytical sedimentation data for proteins. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (eds Harding, S.E. et al.) pp. 90–125. (The Royal Society of Chemistry, Cambridge, (1992).
Acharya, K.R., Ren, J., Stuart, D.I., Phillips, D.C. & Fenna, R.E. Crystal structure of human α-Lactalbumin at 1.7 Å resolution. J. molec. Biol. 221, 571–581 (1991).
Priestle, J.P. RIBBON: A stereo cartoon drawing program for proteins. J. appl. Crystallogr. 21, 572–576 (1988).
Kauzmann, W. in Sulphur in proteins (Eds Benesch, R. et al.) 93–108 (Academic Press, New York, (1959).
Privalov, P.L. Stability of proteins: proteins which do not present a single cooperative system. Adv. Protein Chem. 35, 1–104 (1982).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Wu, L., Peng, Zy. & Kim, P. Bipartite structure of the α-lactalbumin molten globule. Nat Struct Mol Biol 2, 281–286 (1995). https://doi.org/10.1038/nsb0495-281
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0495-281
This article is cited by
-
Conformational Isomers of Denatured and Unfolded Proteins: Methods of Production and Applications
The Protein Journal (2009)
-
Evidence for barrier-limited protein folding kinetics on the microsecond time scale
Nature Structural Biology (1998)
-
Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis
Nature (1997)
-
A residue-specific NMR view of the non-cooperative unfolding of a molten globule
Natural Structural Biology (1997)
-
Proline scanning mutagenesis of a molten globule reveals non-cooperative formation of a protein's overall topology
Nature Structural & Molecular Biology (1996)