Abstract
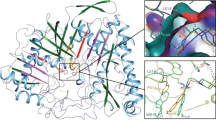
The first crystal structure of a phospholipase D (PLD) family member has been determined at 2.0 Å resolution. The PLD superfamily is defined by a common sequence motif, HxK(x)4D(x)6GSxN, and includes enzymes involved in signal transduction, lipid biosynthesis, endonucleases and open reading frames in pathogenic viruses and bacteria. The crystal structure suggests that residues from two sequence motifs form a single active site. A histidine residue from one motif acts as a nucleophile in the catalytic mechanism, forming a phosphoenzyme intermediate, whereas a histidine residue from the other motif appears to function as a general acid in the cleavage of the phosphodiester bond. The structure suggests that the conserved lysine residues are involved in phosphate binding. Large-scale genomic sequencing revealed that there are many PLD family members. Our results suggest that all of these proteins may possess a common structure and catalytic mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Exton, J.H. Phosphatidylcholine breakdown and signal transduction. Biochim. Biophys. Acta 1212, 26–42 (1994).
Berridge, M.J. Inositol trisphosphate and calcium signalling. Nature 361, 315–325 (1993).
Moolenaar, W.H. Lysophosphatidic acid, a multifunctional phospholipid messenger. J. Biol. Chem. 270, 12949–12952 (1995).
Jalink, K., Hordijk, P.L. & Moolenaar, W.H. Growth factor-like effects of lysophosphatidic acid, a novel lipid mediator. Biochim. Biophys. Acta 1198, 185–196 (1994).
Koonin, E.V. A duplicated catalytic motif in a new superfamily of phosphohydrolases and phospholipid synthases that includes poxvirus envelope proteins. Trends Biochem. Sci. 21, 242–243 (1996).
Ponting, C.P. & Kerr, I.D. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 5, 914–922 (1996).
Zhao, Y., Stuckey, J.A., Lohse, D.L. & Dixon, J.E. Expression, characterization, and crystallization of a member of the novel phospholipase D family of phosphodiesterases. Protein Sci. 6, 2655–2658 (1997).
Pohlman, R.F., Liu, F., Wang, L., More, M.I. & Winans, S.C. Genetic and biochemical analysis of an endonuclease encoded by the IncN plasmid pKM101. Nucleic Acids Res. 21, 4867–4872 (1993).
Gottlin, E.B., Rudolph, A.E., Zhao, Y., Matthews, H.R. & Dixon, J.E. Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate. Proc. Natl. Acad. Sci. USA 95, 9202–9207 (1998).
Van Etten, R.L. & Risley, J.M. Phosphate (oxygen)–water exchange reaction catalyzed by human prostatic acid phosphatase. Proc. Natl. Acad. Sci. USA 75, 4784–4787 (1978).
Rudolph, A.E. et al. Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J. Biol. Chem. in the press (1999).
Raetz, C.R. et al. Phospholipids chiral at phosphorus. Steric course of the reactions catalyzed by phosphatidylserine synthase from Escherichia coli and yeast. Biochemistry 26, 4022–4027 (1987).
Bruzik, K. & Tsai, M.D. Phospholipids chiral at phosphorus. Synthesis of chiral phosphatidylcholine and stereochemistry of phospholipase D. Biochemistry 23, 1656–1661 (1984).
Sung, T.C. et al. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 16, 4519–4530 (1997).
Miller, M., Jaskolski, M., Rao, J.K., Leis, J. & Wlodawer, A. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature 337, 576–579 (1989).
Silva, A.M., Cachau, R.E., Sham, H.L. & Erickson, J.W. Inhibition and catalytic mechanism of HIV-1 aspartic protease. J. Mol. Biol. 255, 321–346 (1996).
Tang, J., James, M.N., Hsu, I.N., Jenkins, J.A. & Blundell, T.L. Structural evidence for gene duplication in the evolution of the acid proteases. Nature 271, 618–621 (1978).
Howard, A.J., Nielsen, C. & Xuong, N.H. Software for a diffractometer with multiwire area detector. Meth. Enz. 114, 452–472 (1985).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enz. 276, 307–326 (1996).
Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Otwinowski, Z. In Isomorphous replacement and anomalous scattering. (eds Wolf, W., Evans, P.R. & Leslie, A.G.W.) 80–86 (Science and Engineering Council/Daresbury Laboratory,Warrington, United Kindom; 1991).
Cowtan, K. DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM newsletter on protein crystallography 31 34–38 (1994).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A. T. X-PLOR Version 3.1: A system for X-ray crystallography and NMR (Yale University Press, New Haven, Connecticut; 1993).
Read R.J. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42, 140–149 (1986).
Cherepanov, P.A. et al. Cloning and detailed mapping of the fra-ymt region of the Yersinia pestis pFra plasmid]. Mol. Gen. Mikrobiol. Virusol., 19–26 (1991).
Hammond, S.M. et al. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J. Biol. Chem. 270, 29640–29643 (1995).
Kraulis, P.J. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Acknowledgements
We would like to thank M. Ludwig and CHESS for providing synchrotron beam time, and J. Zhou and A. Metzger for technical support. This work was supported by a grant from the National Institutes of Health, NIDDK to J.E.D. and by the Walther Cancer Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Stuckey, J., Dixon, J. Crystal structure of a phospholipase D family member. Nat Struct Mol Biol 6, 278–284 (1999). https://doi.org/10.1038/6716
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/6716
This article is cited by
-
Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C
Nature Communications (2023)
-
The catalytic and structural basis of archaeal glycerophospholipid biosynthesis
Extremophiles (2022)
-
Genome-wide identification and expression analysis of phospholipase D gene in leaves of sorghum in response to abiotic stresses
Physiology and Molecular Biology of Plants (2022)
-
Crystal structure of human PLD1 provides insight into activation by PI(4,5)P2 and RhoA
Nature Chemical Biology (2020)
-
Human PLD structures enable drug design and characterization of isoenzyme selectivity
Nature Chemical Biology (2020)