Abstract
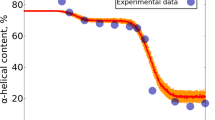
The native state of a protein is generally believed to be the global free energy minimum. However, there is increasing evidence that kinetically selected states play a role in the biological function of some proteins. In a recent folding study of a 125-residue heteropolymer model, one of 200 sequences was found to fold repeatedly to a particular local minimum that did not interconvert to the global minimum. The kinetic preference for this ‘metastable’ state is shown to derive from an entropic barrier associated with inserting a tail segment into the protein interior of the serpin-like global minimum structure. The relation of the present results to the role of metastable states in functioning and pathogenic proteins is discussed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Anfinsen, C.B. Principles that govern the folding of protein chains. Science 181, 223–230 (1973).
Richards, F.M. On the enzymic activity of subtilisin-modified ribonuclease. Proc. Natl. Acad. Sci. USA 44, 162–166 (1958).
Taniuchi, H., Anfinsen, C.B. & Sodja, A., Nuclease-T: An active derivative of staphylococcal nuclease composed of two noncovalently bonded peptide fragments. Proc. Natl. Acad. Sci. USA 58, 1235–1242 (1967).
Taniuchi, H. & Anfinsen, C.B. Simultaneous formation of two alternative enzymically active structures by complementation of two overlapping fragments of staphylococcal nuclease. J. Biol. Chem. 246, 2291–2301 (1971).
Baker, D., Sohl, J.L. & Agard, D.A. A protein-folding reaction under kinetic control.Nature 356, 263–265 (1992).
Hua, Q.-X. et al. Structure of a protein in a kinetic trap. Nature Struct. Biol. 2, 129–137 (1995).
King, J., Haase-Pettingell, C., Robinson, A.S., Speed, M. & Mitraki, A. Thermolabile folding intermediates: Inclusion body precursors and chaperonin substrates. FASEB J. 10, 57–66 (1996).
Carrell, R.W., Evans, D. LI. & Stein, P.E. Mobile reactive center of serpins and the control of thrombosis. Nature 353, 576–578 (1991).
Mottonen, J. et al. Structural basis of latency in plasminogen activator inhibitor-1. Nature 355, 270–273 (1992).
Schulze, A.J., Huber, R., Bode, W. & Engh, R.A. Structural aspects of serpin inhibition. FEBS Lett. 344, 117–124 (1994).
Pan, K.-M. et al. Conversion of α-helices into (β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA 90, 10962–10966 (1993).
Riek, R. et al. NMR structure of the mouse prion protein domain PrP(121-231). Nature 382, 180–182 (1996).
Baker, D. & Agard, D.A. Kinetics versus thermodynamics in protein folding. Biochemistry 33, 7505–7509 (1994).
Karplus, M. & Sali, A. Theories of protein folding. Curr. Opin. Struct. Biol. 5, 58–73 (1995).
Dobson, C.M. Finding the right fold. Nature Struct. Biol. 2, 513–517 (1995).
Dill, K.A. et al. Principles of protein folding—a perspective from simple exact models. Prot. Sci. 4, 561–602 (1995).
Bryngelson, J.D., Onuchic, J.N., Socci, N.D. & Wolynes, P.G., Funnels, pathways, and the energy landscape of protein folding. Proteins 21, 167–195 (1995).
Honeycutt, J.D. & Thirumalai, D. The nature of folded states of globular proteins. Biopolymers 32, 695–709 (1992).
Alonso, D.O.W. & Daggett, V. Molecular dynamics simulations of protein unfolding and limited refolding: Characterization of partially unfolded states of ubiquitin in 60% methanol and water. J. Mol. Biol. 247, 501–520 (1995).
Dinner, A.R., Sali, A. & Karplus, M. The folding mechanism of larger model proteins: Role of native structure. Proc. Natl. Acad. Sci. USA 93, 8356–8361 (1996).
Metropolis, N., Rosenbluth, A.W., Rosenbluth, M.N., Teller, A.H. & Teller, E. Equation of state calculations by fast computing machines. J. Chem. Phys. 21, 1087–1092 (1953).
Baldwin, R.L. On-pathway versus off-pathway folding intermediates. Folding & Design 1, R1–R8 (1996).
Chan, H.S. & Dill, K.A. The effects of internal constraints on the configurations of chain molecules. J. Chem. Phys. 92, 3118–3135 (1990).
Abkevich, V.I., Gutin, A.M. & Shakhnovich, E.I. Specific nucleus as the transition state for protein folding: Evidence from the lattice model. Biochemistry 33, 10026–10036 (1994).
Madras, N. & Sokal, A.D. The pivot algorithm: A highly efficient Monte Carlo method for the self-avoiding walk. J. Stat Phys. 50, 109–186 (1988).
Kauzmann, W. Some factors in the interpretation of protein denaturation. Adv. Prot. Chem. 14, 1–63 (1959).
Kumar, S., Bouzida, D., Swendsen, R.H., Kollman, P.A. & Rosenberg, J.M. The weighted histogram analysis method for free-energy calculations on biomolecules I. The method. J. Comp. Chem. 13, 1011–1021 (1992).
Boczko, E.M. & Brooks, C.L. III . Constant-temperature free energy surfaces for physical and chemical processes. J. Phys. Chem. 97, 4509–4513 (1993).
Fiebig, K.M. & Dill, K.A. Protein core assembly processes. J. Chem. Phys. 98, 3475–3487 (1993).
Dill, K.A., Fiebig, K.M. & Chan, H.S. Cooperativity in protein-folding kinetics. Proc. Natl. Acad. Sci. USA 90, 1942–1946 (1993).
S˘ali, A., Shakhnovich, E. & Karplus, M. Kinetics of protein folding: A lattice model study of the requirements for folding to the native state. J. Mol. Biol. 235, 1614–1636 (1994).
S˘ali, A., Shakhnovich, E. & Karplus, M. How does a protein fold? Nature 369, 248–251 (1994).
Govindarajan, S. & Goldstein, R.A. Searching for foldable protein structures using optimized energy functions. Biopolymers 36, 43–51 (1995).
Govindarajan, S. & Goldstein, R.A. Optimal local propensities for model proteins. Proteins 22, 413–418 (1995).
Abkevich, V.I., Gutin, A.M. & Shakhnovich, E.I. Impact of local and non-local interactions on thermodynamics and kinetics of protein folding. J. Mol. Biol. 252, 460–471 (1995).
Bullough, P.A., Hughson, F.M., Skehel, J.J. & Wiley, D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 (1994).
Zhang, S. & Rich, A. A Direct conversion of an oligopeptide from a β-sheet to an α-helix: A model for amyloid formation. Proc. Natl. Acad. Sci. USA 94, 23–28 (1997).
Booth, D.R. et al. Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature 385, 787–793 (1997).
Perutz, M. Mutations make enzyme polymerize. Nature 385, 773–775 (1997).
Cohen, F.E. et al. Structural clues to prion replication. Science 264, 530–531 (1994).
Nelson, M. et al. MDScope—A visual computing environment for structural biology. Comput. Phys. Commun. 91, 111–134 (1995).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dinner, A., Karplus, M. A metastable state in folding simulations of a protein model. Nat Struct Mol Biol 5, 236–241 (1998). https://doi.org/10.1038/nsb0398-236
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0398-236
This article is cited by
-
Are native proteins metastable?
Nature Chemistry (2011)