Abstract
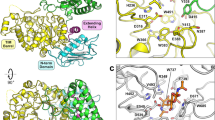
We report the 2.4 Å X-ray crystal structure of a protein with chitosan endo-hydrolase activity isolated from Streptomyces N174. The structure was solved using phases acquired by SIRAS from a two-site methyl mercury derivative combined with solvent flattening and non-crystallographic two-fold symmetry averaging, and refined to an R-factor of 18.5%. The mostly α-helical fold reveals a structural core shared with several classes of lysozyme and barley endochitinase, in spite of a lack of shared sequence. Based on this structural similarity we postulate a putative active site, mechanism of action and mode of substrate recognition. It appears that Glu 22 acts as an acid and Asp 40 serves as a general base to activate a water molecule for an SN2 attack on the glycosidic bond. A series of amino-acid side chains and backbone carbonyl groups may bind the polycationic chitosan substrate in a deep electronegative binding cleft.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Sandford, P. Chitosan: Commercial uses and potential applications. in Chitin and Chitosan (eds Skåjk-Braek, G., Anthonsen, T. & Sandford, P.) 51–69 (Elsevier Applied Science, London, 1989).
Bruck, H.M., Nash, G., Foley, F.D. & Pruiti, B.A., Jr Opportunistic fungal infections of the burn wound with Phycomyces and Aspergillus. Arch. Surgery 102, 476–482 (1971).
Barnicki-Garcia, S. & Nickerson, W.J. Isolation, composition, and structure of cell walls of filamentous and yeast-like forms of. Mucor rouxii Biochim. Biophys. Acta 58, 102–119 (1962).
El Ghaouth, A., Arul, J., Grenier, J. & Asselin, A. Antifungal activity of chitosan on two postharvest pathogens of strawberry fruits. Phytopathology 82, 398–402 (1992).
Hadwiger, L.A., Fristensky, B. & Riggleman, R.C. in Chitin, Chitosan, and Related Enzymes (ed. Zikakis, J. P.) 291–302 (Academic Press, Orlando, 1984).
Davis, B. & Eveleigh, D.E. in Chitin, Chitosan, and Related Enzymes (ed. Zikakis, J. P.) 291–302 (Academic Press, Orlando, 1984).
El Ouakfaoui, S. & Asselin, A. Diversity of chitosanase activity in cucumber. Plant Science 85, 33–41 (1992).
Boucher, I., Dupuy, A., Vidal, P., Neugebauer, W.A. & Brzezinski, R. Purification and characterization of a chitosanase from Streptomyces N174. Appl. Microb. Biotechnol. 38, 188–193 (1992).
Fink, D., Boucher, I., Denis, F. & Brzezinski, R. Cloning and expression in Streptomyces lividans of a chitosanase-encoding gene from the actinomycete Kitasatosporia N174 isolated from soil. Biotech. Lett. 13, 845–850 (1991).
Masson, J.-Y., Denis, F. & Brzezinski, R. Primary sequence of the chitosanase from Streptomyces sp. strain N174 and comparison with other endoglycosidases. Gene 140, 103–107 (1994).
Henrissat, B. & Bairoch, A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293, 781–788 (1993).
Ando, A., Noguchi, K., Yanagi, M., Shinoyama, H., Kagawa, Y., Hirata, H., Yabuki, M. & Fujii, T. Primary structure of chitosanase produced by Bacillus circulans MH-K1. J. Gen. Appl. Microbiol. 38, 135–144 (1992).
Marcotte, E., Hart, P.J., Boucher, I., Brzezinski, R. & Robertus, J.D. Crystallization of a chitosanase from Streptomyces N174. J. Molec. Biol. 232, 995–996 (1993).
Lüthy, R., Bowie, J.U. & Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 356, 83–85 (1992).
Matthews, B.W. & Remington, S.J. The three dimensional structure of 39. the lysozyme from bacteriophage T4. Proc. Natl. Acac. Sci. U.S.A. 71, 4178–4182 (1974).
Rossmann, M.G. & Argos, P. Exploring structural homology of proteins. J. Molec. Biol. 105, 75–96 (1976).
Matthews, B.W., Grütter, M.G., Anderson, W.F. & Remington, S.J. Common precursor of lysozymes of hen egg-white and bacteriophage T4. Nature 290, 334–335 (1981).
Grütter, M.G., Weaver, L.H. & Matthews, B.W. Goose lysozyme structure: an evolutionary link between hen and bacteriophage lysozymes? Nature 303, 828–831 (1983).
Hart, P.J., Monzingo, A.F., Ready, M.P., Ernst, S.R. & Robertus, J.D. Crystal structure of an endochitinase from Hordeum vulgare L. seeds. J. Molec. Biol. 229, 189–193 (1993).
Holm, L. & Sander, C. Structural similarity of plant chitinase and lysozymes from animals and phage. FEBS Lett. 340, 129–132 (1994).
Hart, P.J., Pfluger, H.D., Monzingo, A.F., Hollis, T. & Robertus, J.D. The refined crystal structure of an endochitinase from Hordeum vulgare L. seeds at 1.8 Å resolution. J. Molec. Biol. 248, 402–413 (1995).
Monzingo, A.F., Marcotte, E.M., Hart, P.J. & Robertus, J.D. Chitinase, chitosanases, and lysozymes can be divided into procaryotic and eukaryotic families sharing a conserved core. Nature Struct. Biol. 3, 133–140 (1996).
Kelly, J.A., Sielecki, A.R., Sykes, BD., James, M.N.G. & Phillips, D.C. X-ray crystallography of the binding of the bacterial cell wall trisaccharide NAM-NAG-NAM to lysozyme. Nature 282, 875–878 (1979).
Ford, L.O., Johnson, L.N., Machin, P.A., Phillips, D.C. & Tjian, R. Crystal structure of a lysozyme-tetrasaccharide lactone complex. J. Molec. Biol. 88, 349–371 (1974).
Blake, C.C.F., Koenig, D.F., Mair, G.A., North, A.C.T., Phillips, D.C. & Sarma, V.R. Structure of hen egg-white lysozyme. Nature 206, 757–761 (1965).
Imoto, I., Johnson, L.N., Machin, P.A., Phillips, D.C. & Rupley, J.A. in The Enzymes Vol. 7 (ed. Boyer, P.) 665–868 (Academic Press, New York, 1972).
Matthews, B.W., Remington, S.J., Grütter, M.G. & Anderson, W.F. Relation between hen egg white lysozyme and bacteriophage T4 lysozyme: Evolutionary implications. J. Molec. Biol. 147, 545–558 (1981).
Bennet, A.J. & Sinnott, M.L. Complete kinetic isotope effect description of transition states for acid-catalyzed hydrolyses of methyl α- and β-glucopyranosides. J. Am. Chem. Soc. 108, 72–87 (1986).
Amyes, T.L. & Jencks, W.P. Lifetimes of oxocarbenium ions in aqueous solution from common ion inhibition of the solvolysis of α-azido ethers by added azide ion. J. Am. Chem. Soc. 111, 78–88 (1989).
Withers, S.G., Warren, R.A.J., Street, I.P., Rupitz, K., Kempton, J.B. & Aebersold, R. Unequivocal demonstration of the involvement of a glutamate residue as a nucleophile in the mechanism of a “retaining” glycosidase. J. Am. Chem. Soc. 112, 5887–5889 (1990).
Sinnott, M.L. Catalytic mechanisms of enzymic glycosyl transfer. Chem. Rev. 90, 1171–1202 (1990).
Kuroki, R., Weaver, L.H. & Matthews, B.W. A covalent enzyme-substrate intermediate with saccharide distortion in a mutant T4 lysozyme. Science 262, 2030–2033 (1993).
Fukamizo, T., Koga, D. & Goto, S. Comparative biochemistry of chitinases -anomeric form of the reaction products. Biosci. Biotech. Biochem. 59, 311–313 (1995).
Wang, Q., Graham, R.W., Trimbur, D., Warren, R.A.J. & Withers, S.G. Changing enzymatic reaction mechanisms by mutagenesis: conversion of a retaining glucosidase to an inverting enzyme. J. Am. Chem. Soc. 116, 11594–11595 (1994).
McCarter, J.D. & Withers, S.G. Mechanisms of enzymatic glycoside hydrolysis. Curr. Op. Struct. Biol. 4, 885–892 (1994).
Hadfield, A.T. et al. Crystal structure of the mutant D52S hen egg white lysozyme with an oligosaccharide product. J. Molec. Biol. 243, 856–872 (1994).
Weaver, L.H., Grütter, M.G. & Matthews, B.W. The refined structures of goose lysozyme and its complex with a bound trisaccharide show the goose-type lysozymes lack a catalytic aspartate residue. J. Molec. Biol. 245, 54–68 (1995).
Hamlin, R. Multiwire area X-ray diffractometers. Meth. Enzymol. 114, 416–452 (1985).
Xuong, N.H., Nielson, C., Hamlin, R. & Anderson, D. Strategies for data collection from protein crystals using a multiwire counter area detector diffractometer. J. Appl. Crystallogr. 18, 342–350 (1985).
Howard, A.J., Nielsen, C. & Xuong, N.H. Software for a diffractometer with multiwire area detector. Methods Enzymol. 114, 453–472 (1985).
Wang, B.-C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 115, 90–112 (1985).
Jones, T.A. in Computational Crystallography (ed. Sayre, D.) 303–317 (Oxford University, Oxford, 1982).
Bricogne, G. Methods and programs for direct-space exploration of geometric redundancies. Acta Crystallogr. A32, 832–847 (1976).
Tanaka, N. Representation of the fast-rotation function in a polar coordinate system. Acta Crystallogr. A33, 191–193 (1977).
Sussman, J.L. Constrained-restrained least squares (CORELS) refinement of proteins and nucleic acids. Meth. Enzymol. 115, 271–302 (1985).
Brünger, A.T. in Crystallographic Computing 4: Techniques and New Technologies (eds Isaacs, N. W. & Taylor, M. R.) 126–140 (Clarendon Press, Oxford, 1988).
Rutenber, E. et al. Crystallographic refinement of ricin to 2.5 Å. Prot. Struct. Funct. Genet. 10, 240–250 (1991).
Sim, G.A. The distribution of phase angles for structures containing heavy atoms. Acta. Crystallogr. 12, 813–815 (1959).
Monzingo, A.F., Collins, E.J., Ernst, S.R., Irvin, J.D. & Robertus, J.D. The 2. 5 Å structure of pokeweed antiviral protein. J. Molec. Biol. 233, 705–715 (1993).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Prot. Struct. Funct. Genet. 11, 281–293 (1991).
Kraulis, P.J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Marcotte, E., Monzingo, A., Ernst, S. et al. X-ray structure of an anti-fungal chitosanase from streptomyces N174. Nat Struct Mol Biol 3, 155–162 (1996). https://doi.org/10.1038/nsb0296-155
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0296-155
This article is cited by
-
Biochemical characterization and elucidation action mode of a new endolytic chitosanase for efficient preparation of chitosan oligosaccharides
Biomass Conversion and Biorefinery (2023)
-
Purification and biochemical characterization of a novel chitosanase cloned from the gene of Kitasatospora setae KM-6054 and its application in the production of chitooligosaccharides
World Journal of Microbiology and Biotechnology (2023)
-
Insights into promiscuous chitosanases: the known and the unknown
Applied Microbiology and Biotechnology (2022)
-
Identification of a chitosanase from the marine metagenome and its molecular improvement based on evolution data
Applied Microbiology and Biotechnology (2020)
-
Chitinolytic functions in actinobacteria: ecology, enzymes, and evolution
Applied Microbiology and Biotechnology (2018)