Abstract
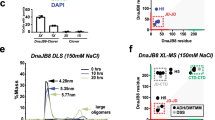
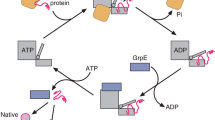
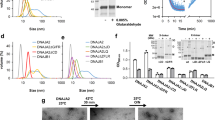
The activity of DnaK (Hsp70) chaperones in assisting protein folding relies on DnaK binding and ATP–controlled release of protein substrates. The ATPase activity of DnaK is tightly controlled by the nucleotide exchange factor GrpE. We find that GrpE interacts stably with the amino–terminal ATPase domain of DnaK. Analysis of the mutant DnaK756 protein, which has a lower affinity for GrpE, reveals a role for residue Gly 32 in GrpE binding. Gly 32 is located in an exposed loop near the nucleotide binding site of DnaK. Deletion of this loop prevents stable GrpE binding, ATPase stimulation by GrpE, and DnaK chaperone activity. Conservation of this loop within the Hsp70 family suggests that cooperation between Hsp70 and GrpE–like proteins may be a general feature of this class of chaperone.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Gething, M.-J. & Sambrook, J. Protein folding in the cell. Nature 355, 33–45 (1992).
Hartl, F.U., Martin, J. & Neupert, W. Protein folding in the cell: the role of molecular chaperones Hsp70 and Hsp60. A. Rev. Biophys. biomol. Struct. 21, 293–322 (1992).
Schröder, H., Langer, T., Hartl, F.U. & Bukau, B. DnaK, DnaJ, GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 12, 4137–4144 (1993).
Skowyra, D., Georgopoulos, C. & Zylicz, M. The E. coli dnaK gene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell 62, 939–944 (1990).
Flynn, G.C., Chappell, T.G. & Rothman, J.E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245, 385–390 (1989).
Flaherty, K.M., Deluca-Flaherty, C. & McKay, D.B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346, 623–628 (1990).
Flaherty, K.M., McKay, D.B. Kabsch W. & Holmes, K.C. Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70K heat shock cognate protein. Proc. natn. Acad. Sci. U.S.A. 88, 5041–5045 (1991).
Bork, P., Sander, C. & Valencia, A., An TPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. natn. Acad. Sci. U.S.A. 89, 7290–7294 (1992).
Holmes, K.C., Sander, C. & Valencia, A. A new ATP-binding fold in actin, hexokinase and Hsc70. Trends Cell Biol. 3, 53–59 (1993).
McCarty, J.S. & Walker, G.C. DnaK as a thermometer: threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc. natn. Acad. Sci. U.S.A. 88, 9513–9517 (1991).
Liberek, K., Marszalek, J., Ang, D., Georgopoulos, C. & Zylicz, M. Escherichia coli. DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. natn. Acad. Sci. U.S.A. 88, 2874–2878 (1991).
Langer, T. et al. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356, 683–689 (1992).
Zylicz, M., Ang, D. & Georgopoulos, C. The grpE protein of Escherichia coli. Purification and properties. J. biol. Chem. 262, 17437–17442(1987).
Georgopoulos, C., Ang, D., Liberek, K. & Zylicz, M. in: Stress proteins in biology and medicine (eds R. Morimoto, A. Tissieres & C. Georgopoulos) 191–222 (Cold Spring Harbor Press, New York, 1990).
Gross, C.A., Straus, D.B. & Erickson, J.W. in: Stress proteins in biology and medicine (eds R. Morimoto, A. Tissieres & C. Georgopoulos) 167–190 (Cold Spring Harbor Press, New York, 1990).
Yochem, J., Uchida, H. Sunshine, M. Saito, H. Georgopoulos, C. & Feiss, M. Genetic analysis of two genes, dnaJ and dnaK, necessary for Escherichia coli. and bacteriophage lambda DNA replication. Molec. Gen. Genet. 164, 9–14 (1978).
Georgopoulos, C. A new bacterial gene (groPC) which affects lambda DNA replication. Molec. Gen. Genet. 151, 35–39 (1977).
Johnson, C., Chandrasekhar, G.N. & Georgopoulos, C. Escherichia coli. dnaK and grpE heat shock proteins interact both in vivo and in vitro. J. Bact. 171, 1590–1596 (1989).
Miyazaki,T., Tanaka, S. Fujita, H. & Itikawa, H. DNA sequence analysis of the dnaK gene of Escherichia coli B and of two dnaK genes carrying the temperature-sensitive mutations dnaK7(Ts) and dnaK756(Ts). J. Bact. 174, 3715–3722 (1992).
Casadaban, M.J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli. using bacteriophage lambda and Mu. J. molec. Biol. 104, 541–555 (1976).
Stüber, D., Matile, H. & Garotta, G. in: Immunological Methods (eds I. Levkovits & B. Pernis) 121–152 (Academic Press, Orlando, 1990).
Stüber, D. & Bujard, H. Transcription from efficient promotors can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1, 1399–1404 (1982).
Gamer, J., Bujard, H. & Bukau, B. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor sigma 32. Cell 69, 833–842 (1992).
Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Harlow, E. & Lane, D. Antibodies: A Laboratory Manual. (Cold Spring Harbor Press, New York, 1988).
Schmid, F.X. in: Protein Structure: A Practical Approach (ed. IE. Creighton) 283 (IRL, Oxford, 1989).
Bukau, B. & Walker, G. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli. mutants lacking the DnaK chaperone. EMBO J. 9, 4027–4036 (1990).
Miller, J.H. Experiments in molecular genetics. (Cold Spring Harbor Press, New York, 1972).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Buchberger, A., Schröder, H., Büttner, M. et al. A conserved loop in the ATPase domain of the DnaK chaperone is essential for stable binding of GrpE. Nat Struct Mol Biol 1, 95–101 (1994). https://doi.org/10.1038/nsb0294-95
Issue Date:
DOI: https://doi.org/10.1038/nsb0294-95
This article is cited by
-
Protein polarization driven by nucleoid exclusion of DnaK(HSP70)–substrate complexes
Nature Communications (2018)
-
Mechanics of Hsp70 chaperones enables differential interaction with client proteins
Nature Structural & Molecular Biology (2011)
-
Heat, pH Induced Aggregation and Surface Hydrophobicity of S. cerevesiae Ssa1 Protein
The Protein Journal (2010)
-
Molecular chaperone targeting and regulation by BAG family proteins
Nature Cell Biology (2001)
-
Functional antibody production using cell-free translation: Effects of protein disulfide isomerase and chaperones
Nature Biotechnology (1997)