Abstract
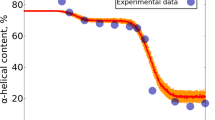
The characterization of unfolded and partly folded states of proteins is central to understanding protein stability and folding, as well as providing a basis for protein design. The four helix bundle–protein interleukin–4 undergoes an unfolding transition at low pH. Using heteronuclear nuclear magnetic resonance methods we show that following this transition the protein retains a highly ordered hydrophobic core in which most, but not all, of the secondary structure is preserved. Extensive disorder exists, however, in regions of polypeptide chain linking the structural elements which make up this core. We suggest that this ‘highly ordered molten globule’ could be indicative of the type of structures occurring late in protein folding processes, in contrast to more disordered ‘molten globules’ which relate to early folding intermediates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Kim, P.S. & Baldwin, R.L. Intermediates in the folding reactions of small proteins. A. Rev. Biochem. 59, 631–660 (1990).
Creighton, T.E. Protein Folding (Freeman, New York, 1992).
Kuwajima, K. The molten globule state as a clue for understanding the folding cooperativity of globular-protein structure. Proteins 6, 87–103 (1989).
Ptitsyn, O.B., Pain, R.H., Semisotnov, G.V., Zerovnik, E & Razgulgaev, O.I. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 262, 20–24 (1990).
Haynie, D.T. & Freire, E. Structural energetics of the molten globule state. Proteins 16, 115–140 (1993).
Dobson, C.M. Unfolded proteins, compact states and molten globules. Curr. Opin. struct. Biol. 2, 6–12 (1992).
Evans, P.A., Dobson, C.M. & Radford, S.E. Probing the structure of folding intermediates. Curr. Opin. struct. Biol. (in the press).
Smith, L.J. et al. Human Interleukin 4: The solution structure of a four-helix-bundle protein. J. molec. Biol. 224, 899–904 (1992).
Powers, R. et al. Three-dimensional solution structure of human interleukin-4 by multidimensional heteronuclear magnetic resonance Spectroscopy. Science 256, 1673–1677 (1992).
Wlodawer, A., Pavlovsky, A. & Gustchina, A. Crystal structure of human recombinant interleukin-4 at 2.25 A resolution. FEBS Lett. 309, 59–64 (1992).
Walter, M.R. et al. Crystal structure of recombinant human interleukin-4. J. biol. Chem. 267, 20371–20376 (1992).
Bazan, J.F. Unravelling the structure of IL-2. Science 257, 410–412 (1992).
Windsor, W.T., Syto, R., Le, H.V. & Trotta, P.P. Analysis of the conformation and stability of Escherichia coli derived recombinant human interleukin 4 by circular dichroism. Biochemistry 30, 1259–1264 (1991).
Dryden, D. & Weir, M.P. Evidence for an acid-induced molten-globule state in interleukin-2; a fluorescence and circular dichroism study. Biochim. biophys. Acta 1078, 94–100 (1991).
Stryer, L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin: A fluorescent probe of non-polar binding sites. J. molec. Biol. 13, 482–495 (1965).
Arakawa, T. & Kenney, W.C. Secondary structure of interleukin-2(Ala125) in unfolded state. Int. J. Peptide Protein Res. 31, 468–473 (1988).
Redfield, C., Boyd, J., Smith, L.J., Smith, R.A.G. & Dobson, C.M. Loop mobility in a four-helix-bundlepProtein: 15N NMR relaxation measurements on human interleukin-4. Biochemistry 31, 10431–10437 (1992).
Kay, L.E., Torchia, D.A. & Bax, A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28, 8972–8979 (1989).
Wüthrich, K. NMR of Proteins and Nucleic Acids (Wiley, New York, 1986).
Dyson, H.J., Rance, M., Houghten, R.A., Wright, P.E. & Lerner, R.A. Folding of immunogenic peptide fragments of proteins in water solution. II. The nascent helix. J. molec. Biol. 201, 201–217 (1988).
Dyson, H.J. & Wright, P.E. Defining solution conformations of small linear peptides. A. Rev. Biophys. biophys. Chem. 20, 519–538 (1991).
Redfield, C. et al. Analysis of the solution structure of human interleukin 4 determined by heteronuclear 3D NMR techniques. submitted to J. molec. Biol.
Pedersen, T.G. et al. A nuclear magnetic resonance study of the hydrogen-exchange behaviour of lysozyme in crystals and solution. J. molec. Biol. 218, 413–426 (1991).
Lee, F. et al. Isolation and characterization of a mouse interleukin cDNA clone that expresses B-cell stimulatory factor 1 activities and T-cell- and mast-cell-stimulating activities. Proc. natn. Acad. Sci. U.S.A. 83, 2061–2065 (1986).
Heussler, T., Eichhorn, M. & Dobbelaere, D.A.F. Cloning of a full-length cDNA encoding bovine interleukin-4 by the polymerase chain reaction. Gene 114, 273–278 (1992).
Diederichs, K., Boone, T. & Karplus, P.A. Novel fold and putative receptor binding site of granulocyte-macrophage colony-stimulating factor. Science 254, 1779–1782 (1991).
DeGrado, W.F., Raleigh, D.P. & Handel, T. De novo protein design: what are we learning? Curr. Opin. struct. Biol. 1, 984–993 (1991).
Brunet, A.P. et al. The role of turns in the structure of an α-helical protein. Nature 364, 355–358 (1993).
Baldwin, R.L. Pulsed H/D-exchange studies of folding intermediates. Curr. Opin. struct. Biol. 3, 84–91 (1993).
Lipari, G. & Szabo, A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. J. Am. chem. Soc. 104, 4546–4570 (1982).
Redfield, C. et al. Secondary structure and topology of human interleukin 4 in solution. Biochemistry 30, 11029–11035 (1991).
Kraulis, P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. appl. Crystallogr. 24, 946–950 (1991).
Wishart, D.S., Sykes, B.D., & Richards, F.M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31, 1647–1651 (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Redfield, C., Smith, R. & Dobson, C. Structural characterization of a highly–ordered ‘molten globule’ at low pH. Nat Struct Mol Biol 1, 23–29 (1994). https://doi.org/10.1038/nsb0194-23
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0194-23
This article is cited by
-
A look back at the molten globule state of proteins: thermodynamic aspects
Biophysical Reviews (2019)
-
Characterization of Molten Globule PopB in Absence and Presence of Its Chaperone PcrH
The Protein Journal (2012)
-
The client protein p53 adopts a molten globule–like state in the presence of Hsp90
Nature Structural & Molecular Biology (2011)
-
Comparative effects of alcohols (methanol, glycerol) and polyethylene glycol (PEG-300) on acid denatured state of goat liver cystatin
Journal of Fluorescence (2011)
-
Conformational Transitions in Ariesaema curvatum Lectin: Characterization of an Acid Induced Active Molten Globule
Journal of Fluorescence (2011)