Key Points
-
Gut microbiota shape immune responses
-
Both innate and adaptive immunity are influenced by gut microbiota, locally in the gut as well as systemically
-
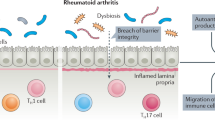
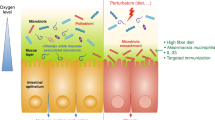
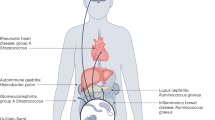
Intestinal dysbiosis is a feature of several inflammatory rheumatic disorders
-
Development of anticitrullinated protein antibodies is modulated by HLA shared epitope alleles, smoking and specialized microbiota at mucosal sites
-
Gut inflammation in spondyloarthritis is linked to a more severe disease course and risk of developing Crohn disease and is associated with intestinal dysbiosis
-
Restoring intestinal homeostasis by altered microbiota is an attractive therapeutic strategy to combat rheumatic diseases
Abstract
The human gut harbours a tremendously diverse and abundant microbial community that correlates with, and even modulates, many health-related processes. The mucosal interfaces are particularly active sites of microorganism–host interplay. Growing insight into the characteristic composition and functionality of the mucosal microbiota has revealed that the microbiota is involved in mucosal barrier integrity and immune function. This involvement affects proinflammatory and anti-inflammatory processes not only at the epithelial level, but also at remote sites such as the joints. Here, we review the role of the gut microbiota in shaping local and systemic immune responses and how disturbances in the host–microorganism interplay can potentially affect the development and progression of rheumatic diseases. Increasing our understanding of how to promote host–microorganism homeostasis could therefore reveal novel strategies for the prevention or alleviation of rheumatic disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Karlsson, F. H., Nookaew, I., Petranovic, D. & Nielsen, J. Prospects for systems biology and modeling of the gut microbiome. Trends Biotechnol. 29, 251–258 (2011).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Moghaddami, M., Cummins, A. & Mayrhofer, G. Lymphocyte-filled villi: comparison with other lymphoid aggregations in the mucosa of the human small intestine. Gastroenterology 115, 1414–1425 (1998).
Pabst, O. et al. Cryptopatches and isolated lymphoid follicles: dynamic lymphoid tissues dispensable for the generation of intraepithelial lymphocytes. Eur. J. Immunol. 35, 98–107 (2005).
Lugering, A. et al. CCR6 identifies lymphoid tissue inducer cells within cryptopatches. Clin. Exp. Immunol. 160, 440–449 (2010).
Claesson, M. J. et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 4, e6669 (2009).
Sankar, S. A., Lagier, J. C., Pontarotti, P., Raoult, D. & Fournier, P. E. The human gut microbiome, a taxonomic conundrum. Syst. Appl. Microbiol. 38, 276–286 (2015).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Ley, R. E. The gene–microbe link. Nature 518, S7 (2015).
Velasquez-Manoff, M. Gut microbiome: the peacekeepers. Nature 518, S3–S11 (2015).
Mirande, C. et al. Dietary fibre degradation and fermentation by two xylanolytic bacteria Bacteroides xylanisolvens XB1AT and Roseburia intestinalis XB6B4 from the human intestine. J. Appl. Microbiol. 109, 451–460 (2010).
Ze, X., Duncan, S. H., Louis, P. & Flint, H. J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543 (2012).
Louis, P. & Flint, H. J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8 (2009).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
Verberkmoes, N. C. et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 3, 179–189 (2009).
Kolmeder, C. A. et al. Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS ONE 7, e29913 (2012).
Koek, M. M., Jellema, R. H., van der Greef, J., Tas, A. C. & Hankemeier, T. Quantitative metabolomics based on gas chromatography mass spectrometry: status and perspectives. Metabolomics 7, 307–328 (2011).
Tuohy, K. M. et al. Studying the human gut microbiota in the trans-omics era — focus on metagenomics and metabonomics. Curr. Pharm. Des. 15, 1415–1427 (2009).
Lin, H. M., Helsby, N. A., Rowan, D. D. & Ferguson, L. R. Using metabolomic analysis to understand inflammatory bowel diseases. Inflamm. Bowel Dis. 17, 1021–1029 (2011).
Helander, H. F. & Fandriks, L. Surface area of the digestive tract — revisited. Scand. J. Gastroenterol. 49, 681–689 (2014).
Delzenne, N. M., Neyrinck, A. M., Backhed, F. & Cani, P. D. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat. Rev. Endocrinol. 7, 639–646 (2011).
Cho, J. H. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 8, 458–466 (2008).
Ogura, Y. et al. Expression of NOD2 in Paneth cells: a possible link to Crohn's ileitis. Gut 52, 1591–1597 (2003).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. USA 107, 14691–14696 (2010).
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009).
Delzenne, N. M. & Cani, P. D. Interaction between obesity and the gut microbiota: relevance in nutrition. Annu. Rev. Nutr. 31, 15–31 (2011).
Algert, C. S., McElduff, A., Morris, J. M. & Roberts, C. L. Perinatal risk factors for early onset of type 1 diabetes in a 2000–2005 birth cohort. Diabet. Med. 26, 1193–1197 (2009).
Thavagnanam, S., Fleming, J., Bromley, A., Shields, M. D. & Cardwell, C. R. A meta-analysis of the association between Caesarean section and childhood asthma. Clin. Exp. Allergy 38, 629–633 (2008).
Decker, E. et al. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Pediatrics 125, e1433–e1440 (2010).
Kull, I., Almqvist, C., Lilja, G., Pershagen, G. & Wickman, M. Breast-feeding reduces the risk of asthma during the first 4 years of life. J. Allergy Clin. Immunol. 114, 755–760 (2004).
Akobeng, A. K., Ramanan, A. V., Buchan, I. & Heller, R. F. Effect of breast feeding on risk of coeliac disease: a systematic review and meta-analysis of observational studies. Arch. Dis. Child. 91, 39–43 (2006).
Shaw, S. Y., Blanchard, J. F. & Bernstein, C. N. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am. J. Gastroenterol. 105, 2687–2692 (2010).
Faria, A. M. & Weiner, H. L. Oral tolerance. Immunol. Rev. 206, 232–259 (2005).
Mowat, A. M. & Agace, W. W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 14, 667–685 (2014).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Fagarasan, S., Kawamoto, S., Kanagawa, O. & Suzuki, K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol. 28, 243–273 (2010).
Tsuji, M. et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity 29, 261–271 (2008).
Fagarasan, S. & Honjo, T. Intestinal IgA synthesis: regulation of front-line body defences. Nat. Rev. Immunol. 3, 63–72 (2003).
Sonnenberg, G. F. & Artis, D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat. Med. 21, 698–708 (2015).
Johansson, M. E. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA 105, 15064–15069 (2008).
Zoetendal, E. G. et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68, 3401–3407 (2002).
Lepage, P. et al. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 11, 473–480 (2005).
Lievin- Le Moal, V. & Servin, A. L. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin. Microbiol. Rev. 19, 315–337 (2006).
Roos, S. & Jonsson, H. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148, 433–442 (2002).
Derrien, M., Vaughan, E. E., Plugge, C. M. & de Vos, W. M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476 (2004).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Eeckhaut, V. et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 62, 1745–1752 (2013).
Khan, M. T. et al. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 6, 1578–1585 (2012).
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O. & Kasper, D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Van Praet, J. T. et al. Commensal microbiota influence systemic autoimmune responses. EMBO J. 34, 466–474 (2015).
Cash, H. L., Whitham, C. V., Behrendt, C. L. & Hooper, L. V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006).
Brandl, K., Plitas, G., Schnabl, B., DeMatteo, R. P. & Pamer, E. G. MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 204, 1891–1900 (2007).
Dessein, R. et al. Toll-like receptor 2 is critical for induction of Reg3β expression and intestinal clearance of Yersinia pseudotuberculosis. Gut 58, 771–776 (2009).
Deshmukh, H. S. et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 20, 524–530 (2014).
Olszak, T. et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012).
Brodin, P. et al. Variation in the human immune system is largely driven by non-heritable influences. Cell 160, 37–47 (2015).
Belkaid, Y. & Segre, J. A. Dialogue between skin microbiota and immunity. Science 346, 954–959 (2014).
Grice, E. A. & Segre, J. A. The skin microbiome. Nat. Rev. Microbiol. 9, 244–253 (2011).
Takemoto, A., Cho, O., Morohoshi, Y., Sugita, T. & Muto, M. Molecular characterization of the skin fungal microbiome in patients with psoriasis. J. Dermatol. 42, 166–170 (2015).
Jagielski, T. et al. Distribution of Malassezia species on the skin of patients with atopic dermatitis, psoriasis, and healthy volunteers assessed by conventional and molecular identification methods. BMC Dermatol. 14, 3 (2014).
Alekseyenko, A. V. et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome 1, 31 (2013).
Statnikov, A. et al. Microbiomic signatures of psoriasis: feasibility and methodology comparison. Sci. Rep. 3, 2620 (2013).
Fahlen, A., Engstrand, L., Baker, B. S., Powles, A. & Fry, L. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch. Dermatol. Res. 304, 15–22 (2012).
Montoya, J. et al. Patients with ankylosing spondylitis have been breast fed less often than healthy controls: a case–control retrospective study. Ann. Rheum. Dis. 75, 879–882 (2016).
Hida, S., Miura, N. N., Adachi, Y. & Ohno, N. Cell wall β-glucan derived from Candida albicans acts as a trigger for autoimmune arthritis in SKG mice. Biol. Pharm. Bull. 30, 1589–1592 (2007).
Baillet, A. C. et al. High Chlamydia burden promotes tumor necrosis factor-dependent reactive arthritis in SKG mice. Arthritis Rheumatol. 67, 1535–1547 (2015).
Benham, H. et al. Interleukin-23 mediates the intestinal response to microbial β-1,3-glucan and the development of spondyloarthritis pathology in SKG mice. Arthritis Rheumatol. 66, 1755–1767 (2014).
Wu, H. J. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Abdollahi-Roodsaz, S. et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Invest. 118, 205–216 (2008).
Ginsburg, I. Can chronic and self-perpetuating arthritis in the human be caused by arthrotropic undegraded microbial cell wall constituants? A working hypothesis. Rheumatol. Rehabil. 16, 141–149 (1977).
Taurog, J. D. et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J. Exp. Med. 180, 2359–2364 (1994).
Khare, S. D., Luthra, H. S. & David, C. S. Spontaneous inflammatory arthritis in HLA-B27 transgenic mice lacking β2-microglobulin: a model of human spondyloarthropathies. J. Exp. Med. 182, 1153–1158 (1995).
Marchesan, J. T. et al. Porphyromonas gingivalis oral infection exacerbates the development and severity of collagen-induced arthritis. Arthritis Res. Ther. 15, R186 (2013).
Dorozynska, I., Majewska-Szczepanik, M., Marcinska, K. & Szczepanik, M. Partial depletion of natural gut flora by antibiotic aggravates collagen induced arthritis (CIA) in mice. Pharmacol. Rep. 66, 250–255 (2014).
Gomez, A. et al. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible *0401 mice but not arthritis-resistant *0402 mice. PLoS ONE 7, e36095 (2012).
Lin, P. et al. HLA-B27 and human β2-microglobulin affect the gut microbiota of transgenic rats. PLoS ONE 9, e105684 (2014).
Hoentjen, F. et al. Reduction of colitis by prebiotics in HLA-B27 transgenic rats is associated with microflora changes and immunomodulation. Inflamm. Bowel Dis. 11, 977–985 (2005).
Rosser, E. C. et al. Regulatory B cells are induced by gut microbiota-driven interleukin-1β and interleukin-6 production. Nat. Med. 20, 1334–1339 (2014).
Vieira, A. T. et al. A role for gut microbiota and the metabolite-sensing receptor GPR43 in a murine model of gout. Arthritis Rheumatol. 67, 1646–1656 (2015).
Mizutani, A. et al. Pristane-induced autoimmunity in germ-free mice. Clin. Immunol. 114, 110–118 (2005).
Maldonado, M. A. et al. The role of environmental antigens in the spontaneous development of autoimmunity in MRL-lpr mice. J. Immunol. 162, 6322–6330 (1999).
East, J., Prosser, P. R., Holborow, E. J. & Jaquet, H. Autoimmune reactions and virus-like particles in germ-free NZB mice. Lancet 1, 755–757 (1967).
Fernandes, G., Friend, P., Yunis, E. J. & Good, R. A. Influence of dietary restriction on immunologic function and renal disease in (NZB × NZW) F1 mice. Proc. Natl Acad. Sci. USA 75, 1500–1504 (1978).
Hsieh, C. C. & Lin, B. F. Dietary factors regulate cytokines in murine models of systemic lupus erythematosus. Autoimmun. Rev. 11, 22–27 (2011).
Liao, X. et al. Paradoxical effects of all-trans-retinoic acid on lupus-like disease in the MRL/lpr mouse model. PLoS ONE 10, e0118176 (2015).
Johnson, B. M., Gaudreau, M. C., Al-Gadban, M. M., Gudi, R. & Vasu, C. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin. Exp. Immunol. 181, 323–337 (2015).
Palm, O., Moum, B., Jahnsen, J. & Gran, J. T. Fibromyalgia and chronic widespread pain in patients with inflammatory bowel disease: a cross sectional population survey. J. Rheumatol. 28, 590–594 (2001).
Salvarani, C. et al. Musculoskeletal manifestations in a population-based cohort of inflammatory bowel disease patients. Scand. J. Gastroenterol. 36, 1307–1313 (2001).
Veloso, F. T., Carvalho, J. & Magro, F. Immune-related systemic manifestations of inflammatory bowel disease. A prospective study of 792 patients. J. Clin. Gastroenterol. 23, 29–34 (1996).
Ditisheim, S. et al. Inflammatory articular disease in patients with inflammatory bowel disease: result of the Swiss IBD Cohort Study. Inflamm. Bowel Dis. 21, 2598–2604 (2015).
Orchard, T. R. et al. The prevalence, clinical features and association of HLA-B27 in sacroiliitis associated with established Crohn's disease. Aliment. Pharmacol. Ther. 29, 193–197 (2009).
Jacques, P. & Elewaut, D. Joint expedition: linking gut inflammation to arthritis. Mucosal Immunol. 1, 364–371 (2008).
Cuvelier, C. et al. Histopathology of intestinal inflammation related to reactive arthritis. Gut 28, 394–401 (1987).
Mielants, H. et al. The evolution of spondyloarthropathies in relation to gut histology. III. Relation between gut and joint. J. Rheumatol. 22, 2279–2284 (1995).
Van Praet, L. et al. Degree of bone marrow oedema in sacroiliac joints of patients with axial spondyloarthritis is linked to gut inflammation and male sex: results from the GIANT cohort. Ann. Rheum. Dis. 73, 1186–1189 (2014).
Van Praet, L. et al. Microscopic gut inflammation in axial spondyloarthritis: a multiparametric predictive model. Ann. Rheum. Dis. 72, 414–417 (2013).
Nagalingam, N. A. & Lynch, S. V. Role of the microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 18, 968–984 (2012).
Michail, S. et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm. Bowel Dis. 18, 1799–1808 (2012).
Hansen, R. et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am. J. Gastroenterol. 107, 1913–1922 (2012).
Huttenhower, C., Kostic, A. D. & Xavier, R. J. Inflammatory bowel disease as a model for translating the microbiome. Immunity 40, 843–854 (2014).
Segain, J. P. et al. Butyrate inhibits inflammatory responses through NFκB inhibition: implications for Crohn's disease. Gut 47, 397–403 (2000).
Schatteman, L. et al. Gut inflammation in psoriatic arthritis: a prospective ileocolonoscopic study. J. Rheumatol. 22, 680–683 (1995).
Stebbings, S. et al. Comparison of the faecal microflora of patients with ankylosing spondylitis and controls using molecular methods of analysis. Rheumatology (Oxford) 41, 1395–1401 (2002).
Costello, M. E. et al. Intestinal dysbiosis in ankylosing spondylitis. Arthritis Rheumatol. http://dx.doi.org/10.1002/art.38967 (2014).
Stoll, M. L. et al. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis Res. Ther. 16, 486 (2014).
Stebbings, S. M., Taylor, C., Tannock, G. W., Baird, M. A. & Highton, J. The immune response to autologous bacteroides in ankylosing spondylitis is characterized by reduced interleukin 10 production. J. Rheumatol. 36, 797–800 (2009).
Gao, Z., Tseng, C. H., Strober, B. E., Pei, Z. & Blaser, M. J. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE 3, e2719 (2008).
Eppinga, H., Konstantinov, S. R., Peppelenbosch, M. P. & Thio, H. B. The microbiome and psoriatic arthritis. Curr. Rheumatol. Rep. 16, 407 (2014).
Scher, J. U. et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 67, 128–139 (2015).
Manichanh, C. et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211 (2006).
McInnes, I. B. & Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 365, 2205–2219 (2011).
Wolff, B. et al. Oral status in patients with early rheumatoid arthritis: a prospective, case–control study. Rheumatology (Oxford) 53, 526–531 (2014).
Scher, J. U. et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 64, 3083–3094 (2012).
Al-Katma, M. K., Bissada, N. F., Bordeaux, J. M., Sue, J. & Askari, A. D. Control of periodontal infection reduces the severity of active rheumatoid arthritis. J. Clin. Rheumatol. 13, 134–137 (2007).
McGraw, W. T., Potempa, J., Farley, D. & Travis, J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect. Immun. 67, 3248–3256 (1999).
Rosenstein, E. D., Greenwald, R. A., Kushner, L. J. & Weissmann, G. Hypothesis: the humoral immune response to oral bacteria provides a stimulus for the development of rheumatoid arthritis. Inflammation 28, 311–318 (2004).
Liao, F. et al. Porphyromonas gingivalis may play an important role in the pathogenesis of periodontitis-associated rheumatoid arthritis. Med. Hypotheses 72, 732–735 (2009).
Wegner, N. et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 62, 2662–2672 (2010).
Kinloch, A. J. et al. Immunization with Porphyromonas gingivalis enolase induces autoimmunity to mammalian α-enolase and arthritis in DR4-IE-transgenic mice. Arthritis Rheum. 63, 3818–3823 (2011).
Hitchon, C. A. et al. Antibodies to Porphyromonas gingivalis are associated with anticitrullinated protein antibodies in patients with rheumatoid arthritis and their relatives. J. Rheumatol. 37, 1105–1112 (2010).
Okada, M. et al. Periodontal treatment decreases levels of antibodies to Porphyromonas gingivalis and citrulline in patients with rheumatoid arthritis and periodontitis. J. Periodontol. 84, e74–e84 (2013).
Martinez-Martinez, R. E. et al. Detection of periodontal bacterial DNA in serum and synovial fluid in refractory rheumatoid arthritis patients. J. Clin. Periodontol. 36, 1004–1010 (2009).
Vaahtovuo, J., Munukka, E., Korkeamaki, M., Luukkainen, R. & Toivanen, P. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 35, 1500–1505 (2008).
Zhang, X. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905 (2015).
Scher, J. U. et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013).
Cuchacovich, R. et al. Detection of bacterial DNA in Latin American patients with reactive arthritis by polymerase chain reaction and sequencing analysis. J. Rheumatol. 29, 1426–1429 (2002).
Morris, A. et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 187, 1067–1075 (2013).
Erb-Downward, J. R. et al. Analysis of the lung microbiome in the 'healthy' smoker and in COPD. PLoS ONE 6, e16384 (2011).
Hilty, M. et al. Disordered microbial communities in asthmatic airways. PLoS ONE 5, e8578 (2010).
Segal, L. N. et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1, 19 (2013).
Makrygiannakis, D. et al. Smoking increases peptidylarginine deiminase 2 enzyme expression in human lungs and increases citrullination in BAL cells. Ann. Rheum. Dis. 67, 1488–1492 (2008).
Gan, R. W. et al. Relationship between air pollution and positivity of RA-related autoantibodies in individuals without established RA: a report on SERA. Ann. Rheum. Dis. 72, 2002–2005 (2013).
Willis, V. C. et al. Sputum autoantibodies in patients with established rheumatoid arthritis and subjects at risk of future clinically apparent disease. Arthritis Rheum. 65, 2545–2554 (2013).
Lugli, E. B. et al. Expression of citrulline and homocitrulline residues in the lungs of non-smokers and smokers: implications for autoimmunity in rheumatoid arthritis. Arthritis Res. Ther. 17, 9 (2015).
Hevia, A. et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio 5, e01548–01514 (2014).
Castelino, M., Eyre, S., Upton, M., Ho, P. & Barton, A. The bacterial skin microbiome in psoriatic arthritis, an unexplored link in pathogenesis: challenges and opportunities offered by recent technological advances. Rheumatology (Oxford) 53, 777–784 (2014).
Arbuckle, M. R. et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N. Engl. J. Med. 349, 1526–1533 (2003).
Ghouri, Y. A. et al. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin. Exp. Gastroenterol. 7, 473–487 (2014).
Kerman, D. H. & Deshpande, A. R. Gut microbiota and inflammatory bowel disease: the role of antibiotics in disease management. Postgrad. Med. 126, 7–19 (2014).
Van Praet, L., Jacques, P., Van den Bosch, F. & Elewaut, D. The transition of acute to chronic bowel inflammation in spondyloarthritis. Nat. Rev. Rheumatol. 8, 288–295 (2012).
Kruglov, A. A. et al. Nonredundant function of soluble LTα3 produced by innate lymphoid cells in intestinal homeostasis. Science 342, 1243–1246 (2013).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Kokkonen, H. et al. Associations of antibodies against citrullinated peptides with human leukocyte antigen-shared epitope and smoking prior to the development of rheumatoid arthritis. Arthritis Res. Ther. 17, 125 (2015).
Too, C. L. et al. Smoking interacts with HLA-DRB1 shared epitope in the development of anti-citrullinated protein antibody-positive rheumatoid arthritis: results from the Malaysian Epidemiological Investigation of Rheumatoid Arthritis (MyEIRA). Arthritis Res. Ther. 14, R89 (2012).
Mahdi, H. et al. Specific interaction between genotype, smoking and autoimmunity to citrullinated α-enolase in the etiology of rheumatoid arthritis. Nat. Genet. 41, 1319–1324 (2009).
Lundstrom, E., Kallberg, H., Alfredsson, L., Klareskog, L. & Padyukov, L. Gene–environment interaction between the DRB1 shared epitope and smoking in the risk of anti-citrullinated protein antibody-positive rheumatoid arthritis: all alleles are important. Arthritis Rheum. 60, 1597–1603 (2009).
Kallberg, H. et al. Gene–gene and gene–environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am. J. Hum. Genet. 80, 867–875 (2007).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Chassaing, B. et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 (2015).
Evans, C. H., Ghivizzani, S. C. & Robbins, P. D. Gene therapy of the rheumatic diseases: 1998 to 2008. Arthritis Res. Ther. 11, 209 (2009).
Edwards, C. J. & Cooper, C. Early environmental factors and rheumatoid arthritis. Clin. Exp. Immunol. 143, 1–5 (2006).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Gibson, S. A., McFarlan, C., Hay, S. & MacFarlane, G. T. Significance of microflora in proteolysis in the colon. Appl. Environ. Microbiol. 55, 679–683 (1989).
Marzorati, M. et al. In vitro modulation of the human gastrointestinal microbial community by plant-derived polysaccharide-rich dietary supplements. Int. J. Food Microbiol. 139, 168–176 (2010).
Voreades, N., Kozil, A. & Weir, T. L. Diet and the development of the human intestinal microbiome. Front. Microbiol. 5, 494 (2014).
Saad, N., Delattre, C., Urdaci, M., Schmitter, J. M. & Bressollier, P. An overview of the last advances in probiotic and prebiotic field. Lebenson. Wiss. Technol. 50, 1–16 (2013).
Preidis, G. A. & Versalovic, J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology 136, 2015–2031 (2009).
Roberfroid, M. et al. Prebiotic effects: metabolic and health benefits. Br. J. Nutr. 104, S1–S63 (2010).
Neyrinck, A. M. et al. Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, Roseburia and Bacteroides/Prevotella in diet-induced obese mice. PLoS ONE 6, e20944 (2011).
Everard, A. et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 60, 2775–2786 (2011).
Van Loo, J. et al. Functional food properties of non-digestible oligosaccharides: a consensus report from the ENDO project (DGXII AIRII-CT94-1095). Br. J. Nutr. 81, 121–132 (1999).
Alipour, B. et al. Effects of Lactobacillus casei supplementation on disease activity and inflammatory cytokines in rheumatoid arthritis patients: a randomized double-blind clinical trial. Int. J. Rheum. Dis. 17, 519–527 (2014).
Vaghef-Mehrabany, E. et al. Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis. Nutrition 30, 430–435 (2014).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
McFarland, L. V. Use of probiotics to correct dysbiosis of normal microbiota following disease or disruptive events: a systematic review. BMJ Open 4, e005047 (2014).
Lemon, K. P., Armitage, G. C., Relman, D. A. & Fischbach, M. A. Microbiota-targeted therapies: an ecological perspective. Sci. Transl. Med. 4, 137rv5 (2012).
Khoruts, A., Dicksved, J., Jansson, J. K. & Sadowsky, M. J. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J. Clin. Gastroenterol. 44, 354–360 (2010).
de Vrieze, J. Medical research. The promise of poop. Science 341, 954–957 (2013).
Allen-Vercoe, E. et al. A Canadian Working Group report on fecal microbial therapy: microbial ecosystems therapeutics. Can. J. Gastroenterol. 26, 457–462 (2012).
Petrof, E. O. et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: 'RePOOPulating' the gut. Microbiome 1, 3 (2013).
Dziechciarz, P., Horvath, A., Shamir, R. & Szajewska, H. Meta-analysis: enteral nutrition in active Crohn's disease in children. Aliment. Pharmacol. Ther. 26, 795–806 (2007).
Holmes, E. et al. Therapeutic modulation of microbiota–host metabolic interactions. Sci. Transl. Med. 4, 137rv6 (2012).
Sonnenburg, J. L. Microbiome engineering. Nature 518, S10 (2015).
Donaldson, G. P., Lee, S. M. & Mazmanian, S. K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016).
Ottman, N., Smidt, H., de Vos, W. M. & Belzer, C. The function of our microbiota: who is out there and what do they do? Front. Cell. Infect. Microbiol. 2, 104 (2012).
Sekirov, I., Russell, S. L., Antunes, L. C. & Finlay, B. B. Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 (2010).
Aziz, Q., Dore, J., Emmanuel, A., Guarner, F. & Quigley, E. M. Gut microbiota and gastrointestinal health: current concepts and future directions. Neurogastroenterol. Motil. 25, 4–15 (2013).
Lawley, T. D. & Walker, A. W. Intestinal colonization resistance. Immunology 138, 1–11 (2013).
Delzenne, N. M. & Cani, P. D. A place for dietary fibre in the management of the metabolic syndrome. Curr. Opin. Clin. Nutr. Metab. Care 8, 636–640 (2005).
Schmidt, C. Mental health: thinking from the gut. Nature 518, S12–15 (2015).
Knoop, K. A. & Newberry, R. D. Isolated lymphoid follicles are dynamic reservoirs for the induction of intestinal IgA. Front. Immunol. 3, 84 (2012).
Author information
Authors and Affiliations
Contributions
T.V.d.W., J.T.V.P., M.M., M.B.D. and D.E. researched data for the article. T.V.d.W., J.v.P and D.E. made substantial contributions to the discussion of content. All authors contributed to the writing of the manuscript. T.V.d.W., J.v.P., M.B.D. and D.E., reviewed/edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Van de Wiele, T., Van Praet, J., Marzorati, M. et al. How the microbiota shapes rheumatic diseases. Nat Rev Rheumatol 12, 398–411 (2016). https://doi.org/10.1038/nrrheum.2016.85
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrrheum.2016.85
This article is cited by
-
Association between primary Sjögren’s syndrome and gut microbiota disruption: a systematic review and meta-analysis
Clinical Rheumatology (2024)
-
The star target in SLE: IL-17
Inflammation Research (2023)
-
The effect of probiotic cheese consumption on inflammatory and anti-inflammatory markers, disease severity, and symptoms in patients with rheumatoid arthritis: study protocol for a randomized, double-blind, placebo-controlled trial
Trials (2022)
-
In vitro triple coculture with gut microbiota from spondyloarthritis patients is characterized by inter-individual differences in inflammatory responses
Scientific Reports (2022)
-
Lycium barbarum polysaccharide modulates gut microbiota to alleviate rheumatoid arthritis in a rat model
npj Science of Food (2022)