Abstract
Genetic studies have revealed that most loci associated with osteoarthritis (OA) show ethnic stratification, with limited overlap between Asian and European populations. Consequently, such studies have often focused on particular ethnic groups, with those performed in European cohorts yielding the most replicated associations. As for other common diseases, the OA susceptibility loci mapped to date account for only a fraction of disease heritability. Nevertheless, analysis of these loci could identify biological pathways related to OA pathogenesis. Such an approach is taken in this Review and provides valuable insights into OA aetiology. For example, several of the loci associated with OA contain genes encoding key regulators of skeletogenesis and endochondral ossification. Furthermore, direct and indirect regulation of gene transcription is highlighted as an important factor in this disease. Interestingly, genes encoding structural proteins of the cartilage extracellular matrix do not seem to be a repository for OA susceptibility. Therefore, susceptibility might operate at a regulatory rather than a structural level, which is a potentially promising finding, as the activities of regulators are amenable to therapeutic modulation. Greater clarity will emerge as more association signals are identified; nonetheless, patterns of aetiology are clearly discernible, from a molecular perspective, even with the relatively small number currently available.
Key Points
-
The currently available data suggest that genetic risk of osteoarthritis (OA) manifests through multiple biological pathways
-
Several core pathways are implicated in OA pathogenesis, in particular those mediating skeletogenesis, encompassing development and differentiation of osteoblasts and chondrocytes
-
Genes encoding structural proteins do not seem to be major contributors to OA susceptibility
-
Transcriptional regulation is a key process associated with OA risk; several susceptibility genes encode either direct transcriptional regulators or indirect regulators that operate via cellular signalling pathways
-
Such regulators are potentially amenable to modulation, more so than structural proteins, offering the possibility of exploiting genetic advances for the development of novel therapeutics
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Reynard, L. N. & Loughlin, J. The genetics and functional analysis of primary osteoarthritis susceptibility. Expert Rev. Mol. Med. 15, e2 (2013).
Jackson, G. C. et al. Pseudoachondroplasia and multiple epiphyseal dysplasia: a 7-year comprehensive analysis of the known disease genes identify novel and recurrent mutations and provides an accurate assessment of their relative contribution. Hum. Mutat. 33, 144–157 (2012).
arcOGEN Consortium and arcOGEN Collaborators. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 380, 815–823 (2012).
Yasuda, T. et al. Aberrations of 6q13 mapped to the COL12A1 locus in chondromyxoid fibroma. Mod. Pathol. 22, 1499–1506 (2009).
Panoutsopoulou K. et al. Insights into the genetic architecture of osteoarthritis from stage 1 of the arcOGEN study. Ann. Rheum. Dis. 70, 864–867 (2011).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
National Institute of Allergy and Infectious Disease (NIAID), NIH. DAVID Bioinformatics Resource 6.7 [online], (2010).
Mackie, E. J., Tatarczuch, L. & Mirams, M. The skeleton: a multi-functional complex organ: the growth plate chondrocyte and endochondral ossification. J. Endocrinol. 211, 109–121 (2011).
Dreier, R. Hypertrophic differentiation of chondrocytes in osteoarthritis: the developmental aspect of degenerative joint disorders. Arthritis Res. Ther. 12, 216 (2010).
Kerkhof, H. J. et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum. 62, 499–510 (2010).
Day-Williams, A. G. et al. A variant in MCF2L is associated with osteoarthritis. Am. J. Hum. Genet. 89, 446–450 (2011).
Chapman, K. et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5′UTR of GDF5 with osteoarthritis susceptibility. Hum. Mol. Genet. 17, 1497–1504 (2008).
Geyer, M. et al. Differential transcriptome analysis of intraarticular lesional vs intact cartilage reveals new candidate genes on osteoarthritis pathophysiology. Osteoarthritis Cartilage 17, 328–335 (2009).
Karlsson, C. et al. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage 18, 581–592 (2010).
Xu, Y. et al. Identification of the pathogenic pathways in osteoarthritic hip cartilage: commonality and discord between hip and knee OA. Osteoarthritis Cartilage 20, 1029–1038 (2012).
Leijten, J. C. et al. Gremlin 1, frizzled-related protein, and Dkk-1 are key regulators of human articular cartilage homeostasis. Arthritis Rheum. 64, 3302–3312 (2012).
Provot, S. & Schipani, E. Molecular mechanisms of endochondral bone development. Biochem. Biophys. Res. Commun. 328, 658–665 (2005).
Klüppel, M. et al. Maintenance of chondroitin sulfation balance by chondroitin-4-sulfotransferase 1 is required for chondrocyte development and growth factor signaling during cartilage morphogenesis. Development 132, 3989–4003 (2005).
Guo, J. et al. PTH/PTHrP receptor delays chondrocyte hypertrophy via both Runx2-dependent and -independent pathways. Dev. Biol. 292, 116–128 (2006).
Kamekura, S. et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 54, 2462–2470 (2006).
Hirata, M. et al. C/EBPβ and RUNX2 cooperate to degrade cartilage with MMP-13 as the target and HIF-2α as the inducer in chondrocytes. Hum. Mol. Genet. 21, 1111–1123 (2012).
Komori, T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 339, 189–195 (2010).
Ueta, C. et al. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J. Cell Biol. 153, 87–100 (2001).
Enomoto, H. et al. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J. Biol. Chem. 275, 8695–8702 (2000).
Fujita, T. et al. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K–Akt signaling. J. Cell Biol. 166, 85–95 (2004).
Kim, I. S. et al. Regulation of chondrocyte differentiation by Cbfa1. Mech. Dev. 80, 159–170 (1999).
Inada, M. et al. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev. Dyn. 214, 279–290 (1999).
Inada, M. et al. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc. Natl Acad. Sci. USA 101, 17192–17197 (2004).
Fosang, A. J. et al. Degradation of cartilage aggrecan by collagenase-3 (MMP-13). FEBS. Lett. 380, 17–20 (1996).
Gelse, K. et al. Molecular differentiation between osteophytic and articular cartilage—clues for a transient and permanent chondrocyte phenotype. Osteoarthritis Cartilage 20, 162–171 (2012).
Vortkamp, A. et al. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273, 613–622 (1996).
Alvarez, J. et al. TGFβ2 mediates the effects of hedgehog on hypertrophic differentiation and PTHrP expression. Development 129, 1913–1924 (2002).
Kronenberg, H. M. PTHrP and skeletal development. Ann. NY Acad. Sci. 1068, 1–13 (2006).
Wysolmerski, J. J. Parathyroid hormone-related protein: an update. J. Clin. Endocrinol. Metab. 97, 2947–2956 (2012).
Mak, K. K., Kronenberg, H. M., Chuang, P. T., Mackem, S. & Yang, Y. Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. Development 135, 1947–1956 (2008).
Lanske, B. et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science 273, 663–666 (1996).
Hirai, T. et al. Parathyroid hormone/parathyroid hormone-related protein receptor signaling is required for maintenance of the growth plate in postnatal life. Proc. Natl Acad. Sci. USA 108, 191–196 (2011).
Li, T. F. et al. Parathyroid hormone-related peptide (PTHrP) inhibits Runx2 expression through the PKA signaling pathway. Exp. Cell Res. 299, 128–136 (2004).
Kozhemyakina, E. et al. Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol. Cell Biol. 29, 5751–5762 (2009).
Zhang, M. et al. PTHrP prevents chondrocyte premature hypertrophy by inducing cyclin-D1-dependent Runx2 and Runx3 phosphorylation, ubiquitylation and proteasomal degradation. J. Cell Sci. 122, 1382–1389 (2009).
Jiang, J. et al. Interaction between zonal populations of articular chondrocytes suppresses chondrocyte mineralization and this process is mediated by PTHrP. Osteoarthritis Cartilage 16, 70–82 (2008).
Kafienah, W. et al. Three-dimensional cartilage tissue engineering using adult stem cells from osteoarthritis patients. Arthritis Rheum. 56, 177–187 (2007).
Petersson, M. et al. Effects of arginine-vasopressin and parathyroid hormone-related protein (1–34) on cell proliferation and production of YKL-40 in cultured chondrocytes from patients with rheumatoid arthritis and osteoarthritis. Osteoarthritis Cartilage 14, 652–659 (2006).
Becher, C. et al. Decrease in the expression of the type 1 PTH/PTHrP receptor (PTH1R) on chondrocytes in animals with osteoarthritis. J. Orthop. Surg. Res. 5, 28 (2010).
Kim, S. Y. & Im, G. I. The expression of SOX trio, PTHrP (parathyroid hormone-related peptide)/IHH (Indian hedgehog protein) in surgically induced osteoarthritis of the rat. Cell Biol. Int. 35, 529–535 (2011).
Serra, R., Karaplis, A. & Sohn, P. Parathyroid hormone-related peptide (PTHrP)-dependent and -independent effects of transforming growth factor beta (TGF-β) on endochondral bone formation. J. Cell Biol. 145, 783–794 (1999).
Chen, C. G. et al. Chondrocyte-intrinsic Smad3 represses Runx2-indicible matrix metalloproteinase 13 expression to maintain articular cartilage and prevent osteoarthritis. Arthritis Rheum. 64, 3278–3289 (2012).
Blaney Davidson, E. N. et al. Increase in ALK1/ALK5 ratio as a cause for elevated MMP-13 expression in osteoarthritis in humans and mice. J. Immunol. 182, 7937–7945 (2009).
Yang, X. et al. TGF-β/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J. Cell Biol. 153, 35–46 (2001).
Wu, Q. et al. Induction of an osteoarthritis-like phenotype and degradation of phosphorylated Smad3 by Smurf2 in transgenic mice. Arthritis Rheum. 58, 3132–3144 (2008).
van de Laar, I. M. et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat. Genet. 43, 121–126 (2011).
Valdes, A. M. et al. Genetic variation in the SMAD3 gene is associated with hip and knee osteoarthritis. Arthritis Rheum. 62, 2347–2352 (2010).
Miyamoto, Y. et al. A functional polymorphism in the 5′-UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat. Genet. 39, 529–533 (2007).
Southam, L. et al. A SNP in the 5′-UTR of GDF5 is associated with osteoarthritis in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum. Mol. Genet. 16, 2226–2232 (2007).
Settle, S. H. et al. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev. Biol. 254, 116–130 (2003).
Tsumaki, N. et al. Role of CDMP-1 in skeletal morphogenesis: promotion of mesenchymal cell recruitment and chondrocyte differentiation. J. Cell Biol. 144, 161–173 (1999).
Coleman, C. M. et al. Growth differentiation factor-5 enhances in vitro mesenchymal stromal cell chondrogenesis and hypertrophy. Stem Cells Dev. 22, 1968–1976 (2013).
Luyten, F. P. Cartilage-derived morphogenetic protein-1. Int. J. Biochem. Cell Biol. 29, 1241–1244 (1997).
Bobacz, K. et al. Differentially regulated expression of growth differentiation factor 5 and bone morphogenetic protein 7 in articular cartilage and synovium in murine chronic arthritis: potential importance for cartilage breakdown and synovial hypertrophy. Arthritis Rheum. 58, 109–118 (2008).
Egli, R. et al. Functional analysis of the osteoarthritis susceptibility-associated GDF5 regulatory polymorphism. Arthritis Rheum. 60, 2055–2064 (2009).
Daans, M., Luyten, F. P. & Lories, R. J. GDF5 deficiency in mice is associated with instability-driven joint damage, gait and subchondral bone changes. Ann. Rheum. Dis. 70, 208–213 (2011).
Okuda, T. et al. Molecular cloning, expression, and chromosomal mapping of human chondroitin 4-sulfotransferase, whose expression pattern in human tissues is different from that of chondroitin 6-sulfotransferase. J. Biochem. 128, 763–770 (2000).
Gandhi, N. S. & Mancera, R. L. The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des. 72, 455–482 (2008).
Klüppel, M. The roles of chondroitin-4-sulfotransferase-1 in development and disease. Prog. Mol. Biol. Transl. Sci. 93, 113–132 (2010).
Cortes, M., Baria, A. T. & Schwartz, N. B. Sulfation of chondroitin sulfate proteoglycans is necessary for proper Indian hedgehog signaling in the developing growth plate. Development 136, 1697–1706 (2009).
Meulenbelt, I. et al. Identification of DIO2 as a new susceptibility locus for symptomatic osteoarthritis. Hum. Mol. Genet. 17, 1867–1875 (2008).
Bos, S. D. et al. Increased type II deiodinase protein in OA-affected cartilage and allelic imbalance of OA risk polymorphism rs225014 at DIO2 in human OA joint tissues. Ann. Rheum. Dis. 71, 1254–1258 (2012).
Shao, Y. Y., Wang, L. & Ballock, R. T. Thyroid hormone and the growth plate. Rev. Endocr. Metab. Disord. 7, 265–271 (2006).
Wang, L., Shao, Y. Y. & Ballock, R. T. Thyroid hormone interacts with the Wnt/β-catenin signaling pathway in the terminal differentiation of growth plate chondrocytes. J. Bone Miner. Res. 22, 1988–1995 (2007).
Williams, A. J. et al. Iodothyronine deiodinase enzyme activities in bone. Bone 43, 126–134 (2008).
Bassett, J. H. et al. Optimal bone strength and mineralization requires the type 2 iodothyronine deiodinase is osteoblasts. Proc. Natl Acad. Sci. USA 107, 7604–7609 (2010).
Nagase, H. et al. Deiodinase 2 upregulation demonstrated in osteoarthritis patients cartilage causes cartilage destruction in tissue-specific transgenic rats. Osteoarthritis Cartilage 21, 514–523 (2013).
Roemer, K. Notch and the p53 clan of transcription factors. Adv. Exp. Med. Biol. 727, 223–240 (2012).
Guerrini, L, Costanzo, A. & Merlo, G. R. A symphony of regulators centered on p63 to control development of ectoderm-derived structures. J. Biomed. Biotechnol. 2011, 864904 (2011).
Barter, M. J., Bui, C. & Young, D. A. Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthritis Cartilage 20, 339–349 (2012).
Nguyen, A. T. & Zhang, Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 25, 1345–1358 (2011).
Castaño Betancourt, M. C. et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc. Natl Acad. Sci. USA 109, 8218–8223 (2012).
Evangelou, E. et al. The DOT1L rs12982744 polymorphism is associated with osteoarthritis of the hip with genome-wide statistical significance in males. Ann. Rheum. Dis. 72, 1264–1265 (2013).
Bos, S. D., Slagboom, P. E. & Meulenbelt, I. New insights into osteoarthritis: early developmental features of an aging-related disease. Curr. Opin. Rheumatol. 20, 553–559 (2008).
Eyre, S. et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 44, 1336–1340 (2012).
Brennan, P., Donev, R. & Hewamana, S. Targeting transcription factors for therapeutic benefit. Mol. Biosyst. 4, 909–919 (2008).
Borgatti, M. et al. Decoy molecules based on PNA-DNA chimeras and targeting Sp1 transcription factors inhibit the activity of urokinase-type plasminogen activator receptor (uPAR) promoter. Oncol. Res. 15, 373–383 (2005).
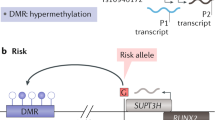
Syddall, C. M., Reynard, L. N., Young, D. A. & Loughlin, J. The identification of trans-acting factors that regulate the expression of GDF5 via the osteoarthritis susceptibility SNP rs143383. PLoS Genet. 9, e1003557 (2013).
Estrada, K. et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 44, 491–501 (2012).
Ermakov, S. et al. Family-based association study of polymorphisms in the RUNX2 locus with hand bone length and hand BMD. Ann. Hum. Genet. 72, 510–518 (2008).
Bustamante, M. et al. Promoter 2 −1025 T/C polymorphism in the RUNX2 gene is associated with femoral neck BMD in Spanish postmenopausal women. Calcif. Tissue Int. 81, 327–332 (2007).
Lee, H. J. et al. Association of a RUNX2 promoter polymorphism with bone mineral density in postmenopausal Korean women. Calcif. Tissue Int. 84, 439–445 (2009).
Plöger, F. et al. Brachydactyly type A2 associated with a defect in proGDF5 processing. Hum. Mol. Genet. 17, 1222–1233 (2008).
Polinkovsky, A. et al. Mutations in CDMP1 cause autosomal dominant brachydactyly type C. Nat. Genet. 17, 18–19 (1997).
Thomas, J. T. et al. A human chondrodysplasia due to a mutation in TGF-β superfamily member. Nat. Genet. 12, 315–317 (1996).
Thomas, J. T. et al. Disruption of human limb morphogenesis by a dominant negative mutation in CDMP1. Nat. Genet. 17, 58–64 (1997).
Szczaluba, K. et al. Du Pan syndrome phenotype caused by heterozygous pathogenic mutations in CDMP1 gene. Am. J. Med. Genet. A. 138, 379–383 (2005).
Dawson, K. et al. GDF5 is a second locus for multiple-synostosis syndrome. Am. J. Hum. Genet. 78, 708–712 (2006).
Klopocki, E. et al. Deletion and point mutations of PTHLH cause brachydactyly type E. Am. J. Hum. Genet. 86, 434–439 (2010).
Celli, J. et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99, 143–153 (1999).
van Bokhoven, H. et al. p63 gene mutations in EEC syndrome, limb-mammary syndrome, and isolated split hand–split foot malformation suggest a genotype–phenotype correlation. Am. J. Hum. Genet. 69, 481–492 (2001).
Ianakiev, P. et al. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am. J. Hum. Genet. 67, 59–66 (2000).
Mundlos, S. et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89, 773–779 (1997).
Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Lettre, G. et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 40, 584–591 (2008).
Zhao, J. et al. The role of height-associated loci identified in genome wide association studies in the determination of pediatric stature. BMC Med. Genet. 11, 96 (2010).
Sanna, S. et al. Common variants in the GDF5–UQCC region are associated with variation in human height. Nat. Genet. 40, 198–203 (2008).
Mendizabal, I, Marigorta, U. M., Lao, O. & Comas, D. Adaptive evolution of loci covarying with the human African Pygmy phenotype. Hum. Genet. 131, 1305–1317 (2012).
Gudbjartsson, D. F. et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 40, 609–615 (2008).
Ermakov, S., Malkin, I., Kobyliansky, E. & Livshits, G. Variation in femoral length is associated with polymorphism in RUNX2 gene. Bone 38, 199–205 (2006).
Kim, J. J. et al. Identification of 15 loci influencing height in a Korean population. J. Hum. Genet. 55, 27–31 (2010).
Elliott, K. S. et al. Evaluation of the genetic overlap between osteoarthritis with body mass index and height using genome-wide association scan data. Ann. Rheum. Dis. 72, 935–941 (2013).
Posthumus, M. et al. The association between the COL12A1 gene and anterior cruciate ligament ruptures. Br. J. Sports Med. 44, 1160–1165 (2010).
Williams, F. M. et al. GDF5 single-nucleotide polymorphism rs143383 is associated with lumbar disc degeneration in Northern European women. Arthritis Rheum. 63, 708–712 (2011).
Rouault, K. et al. Evidence of association between GDF5 polymorphisms and congenital dislocation of the hip in a Caucasian population. Osteoarthritis Cartilage 18, 1144–1149 (2010).
Posthumus, M. et al. Components of the transforming growth factor-β family and the pathogenesis of human Achilles tendon pathology—a genetic association study. Rheumatology (Oxford) 49, 2090–2097 (2010).
Liu, Y. et al. RUNX2 polymorphisms associated with OPLL and OLF in the Han population. Clin. Orthop. Relat. Res. 468, 3333–3341 (2010).
National Center for Biotechnology Information, US National Library of Medicine. Online Mendelian Inheritance in Man (OMIM) [online], (2009).
GWAS Central. GWAS Central 7.0 [online], (2012).
NIH. Genetic Association Database [online], (2011).
National Center for Biotechnology Information, US National Library of Medicine. PubMed [online], (2009).
Acknowledgements
L. Reynard and J. Loughlin acknowledge research support from Arthritis Research UK, the UK Medical Research Council, the JGW Patterson Foundation, the Oliver Bird Rheumatism Program of the Nuffield Foundation, the Dr William Harker Foundation, the NIHR Newcastle Biomedical Research Centre, and the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no 305815 (D-BOARD).
Author information
Authors and Affiliations
Contributions
Both authors researched the data for the article and provided a substantial contribution to discussions of the content and contributed equally to writing the article and to review/editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Reynard, L., Loughlin, J. Insights from human genetic studies into the pathways involved in osteoarthritis. Nat Rev Rheumatol 9, 573–583 (2013). https://doi.org/10.1038/nrrheum.2013.121
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrrheum.2013.121
This article is cited by
-
Inflammaging and Osteoarthritis
Clinical Reviews in Allergy & Immunology (2022)
-
Identification of a potential gene target for osteoarthritis based on bioinformatics analyses
Journal of Orthopaedic Surgery and Research (2020)
-
Gene expression and functional comparison between multipotential stromal cells from lateral and medial condyles of knee osteoarthritis patients
Scientific Reports (2019)
-
Does cartilage ERα overexpression correlate with osteoarthritic chondrosenescence? Indications from Labisia pumila OA mitigation
Journal of Biosciences (2019)
-
An updated meta-analysis of the asporin gene D-repeat in knee osteoarthritis: effects of gender and ethnicity
Journal of Orthopaedic Surgery and Research (2017)