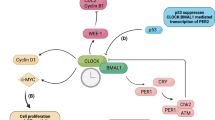
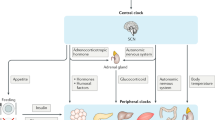
Abstract
The circadian clock machinery is responsible for biological timekeeping on a systemic level. The central clock system controls peripheral clocks through a number of output cues that synchronize the system as a whole. There is growing evidence that changing cellular metabolic states have important effects on circadian rhythms and can thereby influence neuronal function and disease. Epigenetic control has also been implicated in the modulation of biological timekeeping, and cellular metabolism and epigenetic state seem to be closely linked. We discuss the idea that cellular metabolic state and epigenetic mechanisms might work through the circadian clock to regulate neuronal function and influence disease states.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Reppert, S. M. & Weaver, D. R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63, 647–676 (2001).
Bass, J. & Takahashi, J. S. Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010).
Masri, S. & Sassone-Corsi, P. Plasticity and specificity of the circadian epigenome. Nature Neurosci. 13, 1324–1329 (2010).
Eckel-Mahan, K. & Sassone-Corsi, P. Metabolism control by the circadian clock and vice versa. Nature Struct. Mol. Biol. 16, 462–467 (2009).
Green, C. B., Takahashi, J. S. & Bass, J. The meter of metabolism. Cell 134, 728–742 (2008).
Katada, S., Imhof, A. & Sassone-Corsi, P. Connecting threads: epigenetics and metabolism. Cell 148, 24–28 (2012).
Akhtar, R. A. et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12, 540–550 (2002).
Panda, S. et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002).
Thresher, R. J. et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282, 1490–1494 (1998).
Welsh, D. K., Takahashi, J. S. & Kay, S. A. Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577 (2010).
Gallego, M. & Virshup, D. M. Post-translational modifications regulate the ticking of the circadian clock. Nature Rev. Mol. Cell Biol. 8, 139–148 (2007).
Sahar, S. & Sassone-Corsi, P. Metabolism and cancer: the circadian clock connection. Nature Rev. Cancer 9, 886–896 (2009).
Nakahata, Y. et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 (2008).
Asher, G. et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 (2008).
Nakahata, Y., Sahar, S., Astarita, G., Kaluzova, M. & Sassone-Corsi, P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 (2009).
Ramsey, K. M. et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 (2009).
Gerstner, J. R. & Yin, J. C. Circadian rhythms and memory formation. Nature Rev. Neurosci. 11, 577–588 (2010).
Wulff, K., Gatti, S., Wettstein, J. G. & Foster, R. G. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nature Rev. Neurosci. 11, 589–599 (2010).
Huang, W., Ramsey, K. M., Marcheva, B. & Bass, J. Circadian rhythms, sleep, and metabolism. J. Clin. Invest. 121, 2133–2141 (2011).
Yang, C. S. et al. Hypothalamic AMP-activated protein kinase regulates glucose production. Diabetes 59, 2435–2443 (2010).
Oomura, Y., Ono, T., Ooyama, H. & Wayner, M. J. Glucose and osmosensitive neurones of the rat hypothalamus. Nature 222, 282–284 (1969).
Watts, A. G. & Donovan, C. M. Sweet talk in the brain: glucosensing, neural networks, and hypoglycemic counterregulation. Front. Neuroendocrinol. 31, 32–43 (2010).
Trumper, B. G., Reschke, K. & Molling, J. Circadian variation of insulin requirement in insulin dependent diabetes mellitus the relationship between circadian change in insulin demand and diurnal patterns of growth hormone, cortisol and glucagon during euglycemia. Horm. Metab. Res. 27 141–147 (1995).
la Fleur, S. E., Kalsbeek, A., Wortel, J., Fekkes, M. L. & Buijs, R. M. A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes 50, 1237–1243 (2001).
Nagai, K. et al. SCN output drives the autonomic nervous system: with special reference to the autonomic function related to the regulation of glucose metabolism. Prog. Brain Res. 111, 253–272 (1996).
Nagai, K., Nagai, N., Sugahara, K., Niijima, A. & Nakagawa, H. Circadian rhythms and energy metabolism with special reference to the suprachiasmatic nucleus. Neurosci. Biobehav. Rev. 18, 579–584 (1994).
Kalsbeek, A. et al. Circadian control of the daily plasma glucose rhythm: an interplay of GABA and glutamate. PLoS ONE 3, e3194 (2008).
Hirota, T. et al. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J. Biol. Chem. 277, 44244–44251 (2002).
Wang, T. A. et al. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science 337, 839–842 (2012).
Mohawk, J. A., Baer, M. L. & Menaker, M. The methamphetamine-sensitive circadian oscillator does not employ canonical clock genes. Proc. Natl Acad. Sci. USA 106, 3519–3524 (2009).
Pendergast, J. S., Oda, G. A., Niswender, K. D. & Yamazaki, S. Period determination in the food-entrainable and methamphetamine-sensitive circadian oscillator(s). Proc. Natl Acad. Sci. USA 109, 14218–14223 (2012).
Bellet, M. M., Vawter, M. P., Bunney, B. G., Bunney, W. E. & Sassone-Corsi, P. Ketamine influences CLOCK:BMAL1 function leading to altered circadian gene expression. PLoS ONE 6, e23982 (2011).
Jones, C. R. et al. Familial advanced sleep-phase syndrome: a short-period circadian rhythm variant in humans. Nature Med. 5, 1062–1065 (1999).
Toh, K. L. et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040–1043 (2001).
Kyriacou, C. P. & Hastings, M. H. Circadian clocks: genes, sleep, and cognition. Trends Cogn. Sci. 14, 259–267 (2010).
Mignot, E., Taheri, S. & Nishino, S. Sleeping with the hypothalamus: emerging therapeutic targets for sleep disorders. Nature Neurosci. 5, 1071–1075 (2002).
Albrecht, U. Circadian rhythms and sleep the metabolic connection. Pflugers Arch. 463, 23–30 (2011).
Womac, A. D., Burkeen, J. F., Neuendorff, N., Earnest, D. J. & Zoran, M. J. Circadian rhythms of extracellular ATP accumulation in suprachiasmatic nucleus cells and cultured astrocytes. Eur. J. Neurosci. 30, 869–876 (2009).
Asher, G. & Schibler, U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell. Metab. 13, 125–137 (2011).
Kahn, B. B., Alquier, T., Carling, D. & Hardie, D. G. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 1, 15–25 (2005).
Chikahisa, S., Fujiki, N., Kitaoka, K., Shimizu, N. & Sei, H. Central AMPK contributes to sleep homeostasis in mice. Neuropharmacology 57, 369–374 (2009).
Lamia, K. A. et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440 (2009).
Um, J. H. et al. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIɛ)-dependent degradation of clock protein mPer2. J. Biol. Chem. 282, 20794–20798 (2007).
Fulco, M. et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell 14, 661–673 (2008).
Canto, C. et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 (2009).
Dworak, M., McCarley, R. W., Kim, T., Kalinchuk, A. V. & Basheer, R. Sleep and brain energy levels: ATP changes during sleep. J. Neurosci. 30, 9007–9016 (2010).
Zawilska, J. B., Skene, D. J. & Arendt, J. Physiology and pharmacology of melatonin in relation to biological rhythms. Pharmacol. Rep. 61, 383–410 (2009).
Slominski, R. M., Reiter, R. J., Schlabritz-Loutsevitch, N., Ostrom, R. S. & Slominski, A. T. Melatonin membrane receptors in peripheral tissues: distribution and functions. Mol. Cell. Endocrinol. 351, 152–166 (2012).
Arendt, J., Bojkowski, C., Franey, C., Wright, J. & Marks, V. Immunoassay of 6-hydroxymelatonin sulfate in human plasma and urine: abolition of the urinary 24-hour rhythm with atenolol. J. Clin. Endocrinol. Metab. 60, 1166–1173 (1985).
Steindl, P. E. et al. Disruption of the diurnal rhythm of plasma melatonin in cirrhosis. Ann. Intern. Med. 123, 274–277 (1995).
Steindl, P. E., Ferenci, P. & Marktl, W. Impaired hepatic catabolism of melatonin in cirrhosis. Ann. Intern. Med. 127, 494 (1997).
Nishikawa, Y., Shibata, S. & Watanabe, S. Circadian changes in long-term potentiation of rat suprachiasmatic field potentials elicited by optic nerve stimulation in vitro. Brain Res. 695, 158–162 (1995).
Harris, K. M. & Teyler, T. J. Age differences in a circadian influence on hippocampal LTP. Brain Res. 261, 69–73 (1983).
Kondratova, A. A., Dubrovsky, Y. V., Antoch, M. P. & Kondratov, R. V. Circadian clock proteins control adaptation to novel environment and memory formation. Aging 2, 285–297 (2010).
Silva, A. J., Kogan, J. H., Frankland, P. W. & Kida, S. CREB and memory. Annu. Rev. Neurosci. 21, 127–148 (1998).
Gao, J. et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109 (2010).
Alvarez-Saavedra, M. et al. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum. Mol. Genet. 20, 731–751 (2011).
Cheng, H. Y. et al. microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829 (2007).
Michan, S. et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J. Neurosci. 30, 9695–9707 (2010).
Wu, D., Qiu, Y., Gao, X., Yuan, X. B. & Zhai, Q. Overexpression of SIRT1 in mouse forebrain impairs lipid/glucose metabolism and motor function. PLoS ONE 6, e21759 (2011).
Levenson, J. M. et al. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 279, 40545–40559 (2004).
Ding, J. M. et al. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science 266, 1713–1717 (1994).
Doi, M., Hirayama, J. & Sassone-Corsi, P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125, 497–508 (2006).
Ripperger, J. A. & Schibler, U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nature Genet. 38, 369–374 (2006).
Crosio, C., Cermakian, N., Allis, C. D. & Sassone-Corsi, P. Light induces chromatin modification in cells of the mammalian circadian clock. Nature Neurosci. 3, 1241–1247 (2000).
Cheung, P. et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5, 905–915 (2000).
Lo, W. S. et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5, 917–926 (2000).
Gouin, J. P. et al. Altered expression of circadian rhythm genes among individuals with a history of depression. J. Affect. Disord. 126, 161–166 (2010).
Mukherjee, S. et al. Knockdown of Clock in the ventral tegmental area through RNA interference results in a mixed state of mania and depression-like behavior. Biol. Psychiatry 68, 503–511 (2010).
McClung, C. A. Circadian rhythms and mood regulation: insights from pre-clinical models. Eur. Neuropsychopharmacol. 21, S683–S693 (2011).
Doi, M. et al. Impaired light masking in dopamine D2 receptor-null mice. Nature Neurosci. 9, 732–734 (2006).
Hood, S. et al. Endogenous dopamine regulates the rhythm of expression of the clock protein PER2 in the rat dorsal striatum via daily activation of D2 dopamine receptors. J. Neurosci. 30, 14046–14058 (2010).
Yujnovsky, I., Hirayama, J., Doi, M., Borrelli, E. & Sassone-Corsi, P. Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc. Natl Acad. Sci. USA 103, 6386–6391 (2006).
Monteleone, P., Martiadis, V. & Maj, M. Circadian rhythms and treatment implications in depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 35, 1569–1574 (2011).
Golden, R. N. et al. The efficacy of light therapy in the treatment of mood disorders: a review and meta-analysis of the evidence. Am. J. Psychiatry 162, 656–662 (2005).
Ciarleglio, C. M., Axley, J. C., Strauss, B. R., Gamble, K. L. & McMahon, D. G. Perinatal photoperiod imprints the circadian clock. Nature Neurosci. 14, 25–27 (2011).
Roybal, K. et al. Mania-like behavior induced by disruption of CLOCK. Proc. Natl Acad. Sci. USA 104, 6406–6411 (2007).
Rowe, M. K., Wiest, C. & Chuang, D. M. GSK-3 is a viable potential target for therapeutic intervention in bipolar disorder. Neurosci. Biobehav. Rev. 31, 920–931 (2007).
Sahar, S., Zocchi, L., Kinoshita, C., Borrelli, E. & Sassone-Corsi, P. Regulation of BMAL1 protein stability and circadian function by GSK3-mediated phosphorylation. PLoS ONE 5, e8561 (2010).
Yin, L., Wang, J., Klein, P. S. & Lazar, M. A. Nuclear receptor Rev-erbα is a critical lithium-sensitive component of the circadian clock. Science 311, 1002–1005 (2006).
Beaulieu, J. M. et al. Regulation of Akt signaling by D2 and D3 dopamine receptors in vivo. J. Neurosci. 27, 881–885 (2007).
Kohno, T. et al. Effects of lithium on brain glucose metabolism in healthy men. J. Clin. Psychopharmacol. 27, 698–702 (2007).
Borrelli, E., Nestler, E. J., Allis, C. D. & Sassone-Corsi, P. Decoding the epigenetic language of neuronal plasticity. Neuron 60, 961–974 (2008).
Eckel-Mahan, K. L. et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc. Natl Acad. Sci. USA 109, 5541–5546 (2012).
Hatori, M. et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012).
Dallmann, R., Viola, A. U., Tarokh, L., Cajochen, C. & Brown, S. A. The human circadian metabolome. Proc. Natl Acad. Sci. USA 109, 2625–2629 (2012).
Katada, S. & Sassone-Corsi, P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nature Struct. Mol. Biol. 17, 1414–1421 (2010).
Crosio, C., Heitz, E., Allis, C. D., Borrelli, E. & Sassone-Corsi, P. Chromatin remodeling and neuronal response: multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J. Cell Sci. 116, 4905–4914 (2003).
Butcher, G. Q., Lee, B., Cheng, H. Y. & Obrietan, K. Light stimulates MSK1 activation in the suprachiasmatic nucleus via a PACAP-ERK/MAP kinase-dependent mechanism. J. Neurosci. 25, 5305–5313 (2005).
Hirayama, J. et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090 (2007).
Etchegaray, J. P., Lee, C., Wade, P. A. & Reppert, S. M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421, 177–182 (2003).
Curtis, A. M. et al. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 279, 7091–7097 (2004).
Feng, D. et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 (2011).
Alenghat, T. et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456, 997–1000 (2008).
Etchegaray, J. P. et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 281, 21209–21215 (2006).
DiTacchio, L. et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 333, 1881–1885 (2011).
Acknowledgements
We thank all members of the Sassone-Corsi laboratory for helpful discussion. Funding for S.M. was provided by the US National Institutes of Health (NIH) postdoctoral fellowship GM097899. Financial support for P.S.-C was provided bythe US NIH (grant AG041504), INSERM (grant 44790) and Sirtris Pharmaceuticals (SP-48984).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Masri, S., Sassone-Corsi, P. The circadian clock: a framework linking metabolism, epigenetics and neuronal function. Nat Rev Neurosci 14, 69–75 (2013). https://doi.org/10.1038/nrn3393
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrn3393
This article is cited by
-
Sirtuins and the circadian clock interplay in cardioprotection: focus on sirtuin 1
Cellular and Molecular Life Sciences (2021)
-
Circadian and Genetic Modulation of Visually-Guided Navigation in Drosophila Larvae
Scientific Reports (2020)
-
Circadian blueprint of metabolic pathways in the brain
Nature Reviews Neuroscience (2019)
-
Non-coding cis-element of Period2 is essential for maintaining organismal circadian behaviour and body temperature rhythmicity
Nature Communications (2019)
-
The emerging link between cancer, metabolism, and circadian rhythms
Nature Medicine (2018)