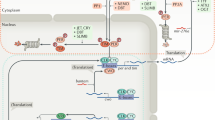
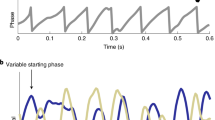
Key Points
-
The nervous system is never silent, and neurons often show daily rhythms in their gene and electrical activities.
-
These rhythms arise from near 24-hour feedback loops that involve identified genes and their protein products.
-
Cellular events that modulate the amplitude and phase of circadian rhythms (including changes in membrane potential and kinase activity) ultimately synchronize circadian cells to each other and to daily environmental cycles.
-
Neurons in several species have been identified as circadian pacemakers for a variety of behaviours, including locomotion, olfaction and learning.
-
All cell types in the body might be capable of intrinsic circadian oscillations.
-
Synaptic strengths change as a function of time of day.
-
The connections between circadian cells and tissues include synaptic and hormonal signals, only some of which have been identified.
-
Daily rhythms in behaviour are normal, and they segregate neural events into sequences that allow the organism to prepare for predictable changes in the environment, such as the availability of sensory cues (for example, light or odour intensity), partners, predators and prey.
Abstract
Biological pacemakers dictate our daily schedules in physiology and behaviour. The molecules, cells and networks that underlie these circadian rhythms can now be monitored using long-term cellular imaging and electrophysiological tools, and initial studies have already suggested a theme — circadian clocks may be crucial for widespread changes in brain activity and plasticity. These daily changes can modify the amount or activity of available genes, transcripts, proteins, ions and other biologically active molecules, ultimately determining cellular properties such as excitability and connectivity. Recently discovered circadian molecules and cells provide preliminary insights into a network that adapts to predictable daily and seasonal changes while remaining robust in the face of other perturbations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dunlap, J. C. Molecular bases for circadian clocks. Cell 96, 271–290 (1999). A wonderful review of the extensive evidence for an intracellular mechanism for circadian rhythm generation that highlights the molecular differences and similarities across species. Although the list of molecular players has grown since this paper was published, it provides an excellent introduction to many of the key concepts in the field of chronobiology.
Hardin, P. E. Essential and expendable features of the circadian timekeeping mechanism. Curr. Opin. Neurobiol. 16, 686–692 (2006).
Debruyne, J. P., Weaver, D. R. & Reppert, S. M. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nature Neurosci. 10, 543–545 (2007).
Hardin, P. E., Hall, J. C. & Rosbash, M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540 (1990). This paper was the first to show that a clock protein suppresses it own transcription, establishing the transcription–translation negative- feedback model for cell-autonomous circadian rhythm generation.
Constance, C. M., Green, C. B., Tei, H. & Block, G. D. Bulla gouldiana period exhibits unique regulation at the mRNA and protein levels. J. Biol. Rhythms 17, 413–427 (2002).
Jeon, M., Gardner, H. F., Miller, E. A., Deshler, J. & Rougvie, A. E. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286, 1141–1146 (1999).
Reppert, S. M. & Weaver, D. R. Coordination of circadian timing in mammals. Nature 418, 935–941 (2002).
Toh, K. L. et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040–1043 (2001). Using an elegant combination of genetics and biochemistry, this paper provided some of the first and best evidence for the molecular genetic basis of sleep–wake rhythms in humans.
Roenneberg, T. & Merrow, M. Circadian clocks — the fall and rise of physiology. Nature Rev. Mol. Cell Biol. 6, 965–971 (2005).
Block, G. D. & McMahon, D. G. Cellular analysis of the Bulla ocular circadian pacemaker system: III. Localization of the circadian pacemaker. J. Comp. Physiol. 155, 387–395 (1984).
Michel, S., Geusz, M. E., Zaritsky, J. J. & Block, G. D. Circadian rhythm in membrane conductance expressed in isolated neurons. Science 259, 239–241 (1993). This study provides the best evidence that single cells in multicellular organisms can act as cell-autonomous circadian pacemakers.
Block, G. D. & Davenport, P. A. Circadian rhythm in Bulla gouldiana: role of the eyes in controlling locomotor behavior. J. Exp. Zoo. 224, 57–63 (1982).
Block, G. D., Geusz, M., Khalsa, S. B. S., Michel, S. & Whitmore, D. Circadian rhythm generation, expression and entrainment in a molluscan model system. Prog. Brain Res. 111, 93–102 (1996).
Helfrich-Forster, C. Techniques that revealed the network of the circadian clock of Drosophila. Methods Enzymol. 393, 439–451 (2005).
Stoleru, D. et al. The Drosophila circadian network is a seasonal timer. Cell 129, 207–219 (2007).
Stoleru, D., Peng, Y., Agosto, J. & Rosbash, M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868 (2004).
Grima, B., Chelot, E., Xia, R. & Rouyer, F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873 (2004).
Lin, Y., Stormo, G. D. & Taghert, P. H. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J. Neurosci. 24, 7951–7957 (2004).
Renn, S. C., Park, J. H., Rosbash, M., Hall, J. C. & Taghert, P. H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802 (1999).
Handler, A. M. & Konopka, R. J. Transplantation of a circadian pacemaker in Drosophila. Nature 279, 236–238 (1979).
Herzog, E. D. & Schwartz, W. J. A neural clockwork for encoding circadian time. J. Appl. Physiol. 92, 401–408 (2002).
Maywood, E. S., O'Neill, J., Wong, G. K., Reddy, A. B. & Hastings, M. H. Circadian timing in health and disease. Prog. Brain Res. 153, 253–269 (2006).
Meyer-Bernstein, E. L. et al. Effects of suprachiasmatic transplants on circadian rhythms of neuroendocrine function in golden hamsters. Endocrinology 140, 207–218 (1999).
Ralph, M. R., Foster, R. G., Davis, F. C. & Menaker, M. Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978 (1990). This now classic paper provided an early demonstration that transplantation of mammalian neurons can rescue circadian behaviour. In this case, transplantation proved that the SCN determines the period of locomotor activity.
Kriegsfeld, L. J., LeSauter, J. & Silver, R. Targeted microlesions reveal novel organization of the hamster suprachiasmatic nucleus. J. Neurosci. 24, 2449–2457 (2004).
Antle, M. C. & Silver, R. Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 28, 145–151 (2005).
Silver, R., LeSauter, J., Tresco, P. A. & Lehman, M. N. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature 382, 810–813 (1996). This elegant transplantation experiment shows that the mammalian clock can regulate locomotor behaviour through a diffusible factor.
Welsh, D. K., Logothetis, D. E., Meister, M. & Reppert, S. M. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706 (1995). This study provides the best evidence that the mammalian SCN is composed of multiple cellular circadian pacemakers.
Yamaguchi, S. et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412 (2003). A technical tour de force in real-time imaging of endogenous gene expression in single cells, this paper indicates that clock cells must spike in order to synchronize to each other and, perhaps, to continue to oscillate.
Deboer, T., Vansteensel, M. J., Detari, L. & Meijer, J. H. Sleep states alter activity of suprachiasmatic nucleus neurons. Nature Neurosci. 6, 1086–1090 (2003).
Gillette, M. U. & Tischkau, S. A. Suprachiasmatic nucleus: the brain's circadian clock. Recent Prog. Horm. Res. 54, 33–58 (1999).
Liu, C. & Reppert, S. M. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron 25, 123–128 (2000).
Aton, S. J., Colwell, C. S., Harmar, A. J., Waschek, J. & Herzog, E. D. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nature Neurosci. 8, 476–483 (2005).
Herzog, E. D. & Huckfeldt, R. M. Circadian entrainment to temperature, but not light, in the isolated suprachiasmatic nucleus. J. Neurophysiol. 90, 763–770 (2003).
Balsalobre, A., Damiola, F. & Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 (1998). This paper dramatically broadened the view of which cell types are capable of circadian oscillations and enabled molecular analyses of rhythm generation in standardized cell culture conditions.
Yu, W. & Hardin, P. E. Circadian oscillators of Drosophila and mammals. J. Cell Sci. 119, 4793–4795 (2006).
Aschoff, J. & Wever, R. A. Human circadian rhythms: a multioscillator system. Fed. Proc. 35, 2326–2332 (1976).
Dijk, D. J. & von Schantz, M. Timing and consolidation of human sleep, wakefulness, and performance by a symphony of oscillators. J. Biol. Rhythms 20, 279–290 (2005).
Plautz, J. D., Kaneko, M., Hall, J. C. & Kay, S. A. Independent photoreceptive circadian clocks throughout Drosophila. Science 278, 1632–1635 (1997).
Krishnan, B., Dryer, S. E. & Hardin, P. E. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature 400, 375–378 (1999). A thorough analysis of the molecular basis of circadian pacemaking function in identified neurons, this paper inspired subsequent studies on the role of the circadian clock in mammalian olfaction.
Zhou, X., Yuan, C. & Guo, A. Drosophila olfactory response rhythms require clock genes but not pigment dispersing factor or lateral neurons. J. Biol. Rhythms 20, 237–244 (2005).
Tosini, G. & Menaker, M. Circadian rhythms in cultured mammalian retina. Science 272, 419–421 (1996).
Granados-Fuentes, D., Saxena, M. T., Prolo, L. M., Aton, S. J. & Herzog, E. D. Olfactory bulb neurons express functional, entrainable circadian rhythms. Eur. J. Neurosci. 19, 898–906 (2004).
Abraham, U., Prior, J. L., Granados-Fuentes, D., Piwnica-Worms, D. R. & Herzog, E. D. Independent circadian oscillations of Period1 in specific brain areas in vivo and in vitro. J. Neurosci. 25, 8620–8626 (2005).
Granados-Fuentes, D., Tseng, A. & Herzog, E. D. A circadian clock in the olfactory bulb controls olfactory responsivity. J. Neurosci. 26, 12219–12225 (2006).
Tataroglu, O., Davidson, A. J., Benvenuto, L. J. & Menaker, M. The methamphetamine-sensitive circadian oscillator (MASCO) in mice. J. Biol. Rhythms 21, 185–194 (2006).
Mieda, M., Williams, S. C., Richardson, J. A., Tanaka, K. & Yanagisawa, M. The dorsomedial hypothalamic nucleus as a putative food-entrainable circadian pacemaker. Proc. Natl Acad. Sci. USA 103, 12150–12155 (2006).
Tosini, G. & Menaker, M. Multioscillatory circadian organization in a vertebrate, Iguana iguana. J. Neurosci. 18, 1105–1114 (1998).
Whitmore, D., Foulkes, N. S. & Sassone-Corsi, P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 404, 87–91 (2000).
Buijs, R. M. et al. Organization of circadian functions: interaction with the body. Prog. Brain Res. 153, 341–360 (2006).
Nagoshi, E. et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004).
Kornmann, B., Schaad, O., Bujard, H., Takahashi, J. S. & Schibler, U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5, e34 (2007).
Abe, M. et al. Circadian rhythms in isolated brain regions. J. Neurosci. 22, 350–356 (2002).
Yoo, S.-H. et al. Period2::luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl Acad. Sci. USA 12, 1–8 (2004).
Yamazaki, S. et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685 (2000). The first use of luciferase as a real-time reporter of circadian gene expression, which provided striking demonstrations of daily rhythms in multiple organs and gave insights into their desynchrony following a shift in the light schedule.
Welsh, D. K., Yoo, S. H., Liu, A. C., Takahashi, J. S. & Kay, S. A. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 14, 2289–2295 (2004).
Carr, A. J. & Whitmore, D. Imaging of single light-responsive clock cells reveals fluctuating free-running periods. Nature Cell Biol. 7, 319–321 (2005).
Reddy, A. B. et al. Circadian orchestration of the hepatic proteome. Curr. Biol. 16, 1107–1115 (2006).
Terazono, H. et al. Adrenergic regulation of clock gene expression in mouse liver. Proc. Natl Acad. Sci. USA 100, 6795–6800 (2003).
Morse, D., Cermakian, N., Brancorsini, S., Parvinen, M. & Sassone-Corsi, P. No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse. Mol. Endocrinol. 17, 141–151 (2003).
Beaver, L. M., Rush, B. L., Gvakharia, B. O. & Giebultowicz, J. M. Noncircadian regulation and function of clock genes period and timeless in oogenesis of Drosophila melanogaster. J. Biol. Rhythms 18, 463–472 (2003).
Wakamatsu, H. et al. Restricted-feeding-induced anticipatory activity rhythm is associated with a phase-shift of the expression of mPer1 and mPer2 mRNA in the cerebral cortex and hippocampus but not in the suprachiasmatic nucleus of mice. Eur. J. Neurosci. 13, 1190–1196 (2001).
Waddington, L. E. et al. Restricted access to food, but not sucrose, saccharine, or salt, synchronizes the expression of Period2 protein in the limbic forebrain. Neuroscience 144, 402–411 (2006).
Stoleru, D., Peng, Y., Nawathean, P. & Rosbash, M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature 438, 238–242 (2005).
Peng, Y., Stoleru, D., Levine, J. D., Hall, J. C. & Rosbash, M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 1, e13 (2003).
Blanchardon, E. et al. Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and period protein overexpression. Eur. J. Neurosci. 13, 871–888 (2001).
Helfrich-Forster, C. et al. Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J. Neurosci. 20, 3339–3353 (2000).
Brown, T., Colwell, C. S., Waschek, J. & Piggins, H. D. Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J. Neurophysiol. 97, 2553–2558 (2006).
Maywood, E. S. et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 16, 599–605 (2006).
Vosko, A. M., Schroeder, A., Loh, D. H. & Colwell, C. S. Vasoactive intestinal peptide and the mammalian circadian system. Gen. Comp. Endocrinol. 152, 165–175 (2007).
Aton, S. J., Huettner, J. E., Straume, M. & Herzog, E. D. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc. Natl Acad. Sci. USA 103, 19188–19193 (2006).
Aton, S. J. & Herzog, E. D. Come together, right... now: synchronization of rhythms in a mammalian circadian clock. Neuron 48, 531–534 (2005).
Nitabach, M. N. Circadian rhythms: clock coordination. Nature 438, 173–175 (2005).
Taghert, P. H. & Shafer, O. T. Mechanisms of clock output in the Drosophila circadian pacemaker system. J. Biol. Rhythms 21, 445–457 (2006).
Page, T. L. & Nalovic, K. G. Properties of mutual coupling between the two circadian pacemakers in the eyes of the mollusc Bulla gouldiana. J. Biol. Rhythms 7, 213–226 (1992).
Yamazaki, S., Kerbeshian, M. C., Hocker, C. G., Block, G. D. & Menaker, M. Rhythmic properties of the hamster suprachiasmatic nucleus in vivo. J. Neurosci. 18, 10709–10723 (1998).
Inouye, S. T. & Kawamura, H. Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc. Natl Acad. Sci. USA 76, 5962–5966 (1979).
Prolo, L. M., Takahashi, J. S. & Herzog, E. D. Circadian rhythm generation and entrainment in astrocytes. J. Neurosci. 25, 404–408 (2005).
Suh, J. & Jackson, F. R. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron 55, 435–447 (2007).
Panda, S. et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002). One of the first reports to show that a significant proportion of the genome is transcribed on a circadian basis. The resulting online database is a useful resource that provides details of tissue-specific and circadian genes (see further information).
Yelamanchili, S. V. et al. Differential sorting of the vesicular glutamate transporter 1 into a defined vesicular pool is regulated by light signaling involving the clock gene Period2. J. Biol. Chem. 281, 15671–15679 (2006).
Kuhlman, S. J. & McMahon, D. G. Encoding the ins and outs of circadian pacemaking. J. Biol. Rhythms 21, 470–481 (2006).
Jackson, A. C., Yao, G. L. & Bean, B. P. Mechanism of spontaneous firing in dorsomedial suprachiasmatic nucleus neurons. J. Neurosci. 24, 7985–7998 (2004).
Pitts, G. R., Ohta, H. & McMahon, D. G. Daily rhythmicity of large-conductance Ca2+-activated K+ currents in suprachiasmatic nucleus neurons. Brain Res. 1071, 54–62 (2006).
Meredith, A. L. et al. BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nature Neurosci. 9, 1041–1049 (2006).
De Jeu, M., Hermes, M. & Pennartz, C. Circadian modulation of membrane properties in slices of rat suprachiasmatic nucleus. Neuroreport 9, 3725–3729 (1998).
Itri, J. N., Michel, S., Vansteensel, M. J., Meijer, J. H. & Colwell, C. S. Fast delayed rectifier potassium current is required for circadian neural activity. Nature Neurosci. 8, 650–656 (2005).
Chaudhury, D., Wang, L. M. & Colwell, C. S. Circadian regulation of hippocampal long-term potentiation. J. Biol. Rhythms 20, 225–236 (2005).
Barnes, C. A., McNaughton, B. L., Goddard, G. V., Douglas, R. M. & Adamec, R. Circadian rhythm of synaptic excitability in rat and monkey central nervous system. Science 197, 91–92 (1977).
Bunning, E. The Physiological Clock: Circadian Rhythms and Biological Chronometry (English Univ. Press, London, 1973).
Inagaki, N., Honma, S., Ono, D., Tanahashi, Y. & Honma, K. I. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc. Natl. Acad. Sci. USA 104, 7664–7669 (2007).
Vanderleest, H. T. et al. Seasonal encoding by the circadian pacemaker of the SCN. Curr. Biol. 17, 468–473 (2007).
Schwartz, W. J., De la Iglesia, H. O., Zlomanczuk, P. & Illnerova, H. Encoding Le Quattro Stagioni within the mammalian brain: photoperiodic ochestration through the suprachiasmatic nucleus. J. Biol. Rhythms 16, 302–311 (2001).
Sandrelli, F. et al. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science 316, 1898–1900 (2007).
Tauber, E. et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science 316, 1895–1898 (2007).
Murad, A., Emery-Le, M. & Emery, P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron 53, 689–701 (2007).
Yoshii, T. et al. Drosophila cryb mutation reveals two circadian clocks that drive locomotor rhythm and have different responsiveness to light. J. Insect. Physiol. 50, 479–488 (2004).
Rieger, D., Shafer, O. T., Tomioka, K. & Helfrich-Forster, C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J. Neurosci. 26, 2531–2543 (2006).
Majercak, J., Sidote, D., Hardin, P. E. & Edery, I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230 (1999).
Janik, D. & Mrosovsky, N. Nonphotically induced phase shifts of circadian rhythms in the golden hamster: activity-response curves at different ambient temperatures. Physiol. Behav. 53, 431–436 (1993).
McArthur, A. J., Gillette, M. U. & Prosser, R. A. Melatonin directly resets the rat suprachiasmatic circadian clock in vitro. Brain Res. 565, 158–161 (1991).
Hall, A. C., Hoffmaster, R. M., Stern, E. L., Harrington, M. E. & Bickar, D. Suprachiasmatic nucleus neurons are glucose sensitive. J. Biol. Rhythms 12, 388–400 (1997).
Nitabach, M. N., Holmes, T. C. & Blau, J. Membranes, ions, and clocks: testing the Njus–Sulzman–Hastings model of the circadian oscillator. Methods Enzymol. 393, 682–693 (2005).
Herzog, E. D., Aton, S. J., Numano, R., Sakaki, Y. & Tei, H. Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J. Biol. Rhythms 19, 35–46 (2004).
Amdaoud, M., Vallade, M., Weiss-Schaber, C. & Mihalcescu, I. Cyanobacterial clock, a stable phase oscillator with negligible intercellular coupling. Proc. Natl Acad. Sci. USA 104, 7051–7056 (2007). A beautiful quantitative analysis of daily rhythms that found no cell–cell circadian communication in this unicellular organism.
Herzog, E. D., Takahashi, J. S. & Block, G. D. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nature Neurosci. 1, 708–713 (1998).
Nakamura, W., Honma, S., Shirakawa, T. & Honma, K.-I. Clock mutation lengthens the circadian period without damping rhythms in individual SCN neurons. Nature Neurosci. 5, 399–400 (2002).
Liu, A. C. et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616 (2007). A provocative and extensive set of experiments that show that the deletion of individual genes can lead to a loss of circadian rhythms in single cells and that cell–cell communication among the mutant cells can rescue rhythmicity.
Gunawan, R. & Doyle, F. J., III. Phase sensitivity analysis of circadian rhythm entrainment. J. Biol. Rhythms 22, 180–194 (2007).
Stelling, J., Gilles, E. D. & Doyle, F. J., III. Robustness properties of circadian clock architectures. Proc. Natl Acad. Sci. USA 101, 13210–13215 (2004).
Ueda, H. R. et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nature Genet. 37, 187–192 (2005). In this study, a wonderful combination of theory and experimentation shows that some events in the circadian transcriptional network are more sensitive to perturbation than others.
Marder, E. & Goaillard, J. M. Variability, compensation and homeostasis in neuron and network function. Nature Rev. Neurosci. 7, 563–574 (2006).
To, T. L., Henson, M. A., Herzog, E. D. & Doyle, F. J., III. A molecular model for intercellular synchronization in the mammalian circadian clock. Biophys. J. 92, 3792–3803 (2007).
Carr, A. J. et al. Photoperiod differentially regulates circadian oscillators in central and peripheral tissues of the Syrian hamster. Curr. Biol. 13, 1543–1548 (2003).
Sauman, I. et al. Connecting the navigational clock to sun compass input in monarch butterfly brain. Neuron 46, 457–467 (2005).
Pittendrigh, C. S. & Daan, S. A functional analysis of circadian pacemakers in nocturnal rodents. V. Pacemaker structure: a clock for all seasons. J. Comp. Physiol. 106, 333–355 (1976). A classic paper that brings together behavioural observations and entrainment theory into an elegant, two-oscillator model of the mammalian circadian system.
Lotze, M., Wittmann, M., von Steinbuchel, N., Poppel, E. & Roenneberg, T. Daily rhythm of temporal resolution in the auditory system. Cortex 35, 89–100 (1999).
Herzog, E. D. & Block, G. D. Keeping an eye on retinal clocks. Chronobiol. Int. 16, 229–247 (1999).
Granados-Fuentes, D., Tseng, A. & Herzog, E. D. A circadian clock in the olfactory bulb controls olfactory responsivity. J. Neurosci. 26, 12219–12225 (2006).
Barlow, R. B. Circadian and efferent modulation of visual sensitivity. Prog. Brain Res. 131, 487–503 (2001).
Stickgold, R. & Walker, M. P. Sleep-dependent memory consolidation and reconsolidation. Sleep Med. 8, 331–343 (2007).
Dijk, D. J. et al. Light treatment for sleep disorders: consensus report. II. Basic properties of circadian physiology and sleep regulation. J. Biol. Rhythms 10, 113–125 (1995).
Wisor, J. P. et al. A role for cryptochromes in sleep regulation. BMC Neurosci. 3, 20 (2002).
Viola, A. U. et al. PER3 polymorphism predicts sleep structure and waking performance. Curr. Biol. 17, 613–618 (2007).
Pace-Schott, E. F. & Hobson, J. A. The neurobiology of sleep: genetics, cellular physiology and subcortical networks. Nature Rev. Neurosci. 3, 591–605 (2002).
Benington, J. H. & Frank, M. G. Cellular and molecular connections between sleep and synaptic plasticity. Prog. Neurobiol. 69, 71–101 (2003).
Cirelli, C., Gutierrez, C. M. & Tononi, G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron 41, 35–43 (2004).
Foster, D. J. & Wilson, M. A. Reverse replay of behavioural sequences in hippocampal place cells during the awake state. Nature 440, 680–683 (2006).
Margoliash, D. Song learning and sleep. Nature Neurosci. 8, 546–548 (2005).
Peigneux, P. et al. Are spatial memories strengthened in the human hippocampus during slow wave sleep? Neuron 44, 535–545 (2004).
Huber, R., Ghilardi, M. F., Massimini, M. & Tononi, G. Local sleep and learning. Nature 430, 78–81 (2004).
Jha, S. K. et al. Sleep-dependent plasticity requires cortical activity. J. Neurosci. 25, 9266–9274 (2005).
Frank, M. G., Issa, N. P. & Stryker, M. P. Sleep enhances plasticity in the developing visual cortex. Neuron 30, 275–287 (2001).
Turek, F. W. et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 (2005).
Levi, F. & Schibler, U. Circadian rhythms: mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 47, 593–628 (2007).
Roybal, K. et al. Mania-like behavior induced by disruption of CLOCK. Proc. Natl Acad. Sci. USA 104, 6406–6411 (2007).
Davidson, A. J. et al. Chronic jet-lag increases mortality in aged mice. Curr. Biol. 16, R914–R916 (2006).
McClung, C. A. Circadian genes, rhythms and the biology of mood disorders. Pharmacol. Ther. 114, 222–232 (2007).
Kripke, D. F., Mullaney, D. J., Atkinson, M. & Wolf, S. Circadian rhythm disorders in manic-depressives. Biol. Psychiatry 13, 335–351 (1978).
Wehr, T. A. et al. Treatment of rapidly cycling bipolar patient by using extended bed rest and darkness to stabilize the timing and duration of sleep. Biol. Psychiatry 43, 822–828 (1998).
Xu, Y. et al. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature 434, 640–644 (2005).
Xu, Y. et al. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell 128, 59–70 (2007).
Welsh, D. K., Imaizumi, T. & Kay, S. A. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 393, 269–288 (2005).
Ikeda, M. et al. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron 38, 253–263 (2003).
Lundkvist, G. B. & Block, G. D. Role of neuronal membrane events in circadian rhythm generation. Methods Enzymol. 393, 623–642 (2005).
Nitabach, M. N., Sheeba, V., Vera, D. A., Blau, J. & Holmes, T. C. Membrane electrical excitability is necessary for the free-running larval Drosophila circadian clock. J. Neurobiol. 62, 1–13 (2004).
Nitabach, M. N., Blau, J. & Holmes, T. C. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109, 485–495 (2002). A technical tour-de-force that shows that chronic hyperpolarization of identified neurons abolishes behavioural and molecular circadian rhythms.
Lundkvist, G. B., Kwak, Y., Davis, E. K., Tei, H. & Block, G. D. A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J. Neurosci. 25, 7682–7686 (2005).
Colwell, C. S. Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur. J. Neurosci. 12, 571–576 (2000).
Barlow, R. B. Jr, Chamberlain, S. C. & Levinson, J. Z. Limulus brain modulates the structure and function of the lateral eyes. Science 210, 1037–1039 (1980).
Acknowledgements
I am deeply grateful to the members of the Washington University Clocksclub (the laboratories of R. Van Gelder, P. Taghert, P. Shaw, L. Muglia and P. Gray) and to M. Hastings, D.-J. Dijk and B. Schwartz for their guidance on this Review. This work was supported by US National Institutes of Health (MH63104 and GM78993) and National Science Foundation (425445) grants.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Glossary
- Clock genes
-
A class of genes, the primary function of which is to participate in a transcription–translation negative-feedback mechanism that generates a near 24-hour rhythm in physiology.
- Cell-autonomous
-
Intrinsic to a single cell. Circadian rhythm generation and output is a property of circadian pacemaking cells.
- Advanced sleep phase syndrome
-
An autosomal-dominant sleep disorder that is characterized by early sleep onset and offset.
- Metazoan
-
An animal belonging to the subkingdom Metazoa, which comprises multicellular animals that have cells that are differentiated into tissues and organs. The Metazon subgroup includes all animals except protozoans and sponges.
- Free-running rhythm
-
An oscillation that persists in the absence of any external input, for example, circadian rhythms in unchanging conditions. When environmental (entraining) cues are eliminated, the intrinsic period of the system is revealed.
- Eclosion
-
The emergence of an adult insect from its pupal case. In many insects, the timing of this emergence is regulated by the circadian clock.
- Malpighian tubules
-
The insect analogue of the kidney. In flies, these excretory tubules express circadian ryhthms in gene expression in vitro and in vivo, even when transplanted into behaviourally arrhythmic flies.
- Real-time reporter
-
A molecular probe that provides information about the level or activity of intracellular events with high temporal resolution compared to the kinetics of the process that is being studied.
- Photoperiodism
-
The biological response to a change in day length. Examples of photoperiodic responses include flowering on long days and storing fat on short days.
- Sensitivity analysis
-
An engineering approach to determine which processes are susceptible to perturbation. The basic strategy is to vary parameters (for example, the maximal transcription rate of the period gene, or the binding rate of the period and cryptochrome proteins) in a model and measure the effect on a specific process (for example, the amplitude or period of circadian oscillations in period proteins). Parameters that cause larger changes in the output when varied over a smaller range are deemed more sensitive.
- Cis regulatory region
-
A segment of DNA physically proximal to a gene that controls the level and timing of that gene's expression. Promoter elements like E-boxes and RREs are found in the cis regulatory regions.
- E box
-
An evolutionarily conserved nucleotide sequence (either CACGTG or CACGTT) in the promoter (cis regulatory) regions of some genes. Through binding of the transcription factors clock and BMAL1, E boxes regulate these genes' circadian expression.
- D box
-
Also known as the DBP/E4BP4 binding element. An evolutionarily conserved nucleotide sequence (TTA[T/C]GTAA, where [T/C] indicates thymidine or cytosine) in the promoter (cis regulatory) regions of some genes. Through binding of specific transcription factors, D boxes regulate these genes' circadian expression.
- RRE
-
Also known as the RevErbA/ROR binding element. An evolutionarily conserved nucleotide sequence ([A/T]A[A/T]NT[A/G]GGTCA, where [A/T] indicates adenosine or thymidine and N indicates any nucleotide) in the promoter (cis regulatory) regions of some genes. Through binding of the transcription factors RevErb and ROR, RREs regulate these genes circadian expression.
Rights and permissions
About this article
Cite this article
Herzog, E. Neurons and networks in daily rhythms. Nat Rev Neurosci 8, 790–802 (2007). https://doi.org/10.1038/nrn2215
Issue Date:
DOI: https://doi.org/10.1038/nrn2215
This article is cited by
-
The intersection between ghrelin, metabolism and circadian rhythms
Nature Reviews Endocrinology (2024)
-
A clock-dependent brake for rhythmic arousal in the dorsomedial hypothalamus
Nature Communications (2023)
-
Theory of localization-hindered thermalization in nonlinear multimode photonics
Communications Physics (2023)
-
A melanopsin ganglion cell subtype forms a dorsal retinal mosaic projecting to the supraoptic nucleus
Nature Communications (2023)
-
The Circadian Clocks, Oscillations of Pain-Related Mediators, and Pain
Cellular and Molecular Neurobiology (2023)