Key Points
-
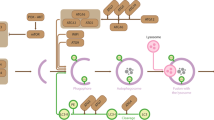
Transcriptional dysregulation is part of the pathogenic mechanism that underlies neuronal dysfunction in polyglutamine repeat diseases such as Huntington's disease (HD). Microarray experiments show that the expression of a subset of genes is robustly altered in mouse models of HD and in the brains of patients with HD.
-
Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are enzymes that control transcription by acetylating and deacetylating histones, thereby changing the conformation of chromatin structure.
-
Expanded polyglutamine repeat proteins adopt aberrant interactions with HATs and HDACs as well as other transcription factors, co-activators and co-repressors owing to conformational changes caused by the polyglutamine stretch within the mutant protein.
-
Of the four classes of HDAC enzyme, class I is ubiquitously expressed and class II is highly expressed in muscle, heart and brain.
-
In addition to deacetylating histones, HDACs also modify non-histone proteins such as the tumour suppressor p53 and heat shock protein 90 (HSP90), both of which are implicated in HD pathogenesis.
-
Compounds that inhibit these class I and II HDACs are in clinical trials for the treatment of many types of cancer. These drugs are currently being tested in preclinical trials using mouse models of polyglutamine repeat disease.
-
One HDAC inhibitor, phenylbutyrate, is in phase II clinical trials for HD, and alterations in blood biomarker expression after treatment look promising.
Abstract
During the past 5 years, gene expression studies in cell culture, animal models and in the brains of patients have shown that the perturbation of transcription frequently results in neuronal dysfunction in polyglutamine repeat diseases such as Huntington's disease. Histone deacetylases act as repressors of transcription through interactions with co-repressor complexes, which leads to chromatin remodelling. Aberrant interactions between polyglutamine proteins and regulators of transcription could be one mechanism by which transcriptional dysregulation occurs. Here, we discuss the potential therapeutic pathways through which histone deacetylase inhibitors might act to correct the aberrant transcription observed in Huntington's disease and other polyglutamine repeat diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bates, G. P., Harper, P. S. & Jones, A. L. (eds) Huntington's Disease (Oxford Univ. Press, Oxford, 2002).
Cummings, C. J. & Zoghbi, H. Y. Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet. 9, 909–916 (2000).
Paulson, H. L. et al. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19, 333–344 (1997).
Davies, S. W. et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90, 537–548 (1997).
DiFiglia, M. et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 (1997).
Wacker, J. L., Zareie, M. H., Fong, H., Sarikaya, M. & Muchowski, P. J. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nature Struct. Mol. Biol. 11, 1215–1222 (2004).
Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004).
Kazantsev, A., Preisinger, E., Dranovsky, A., Goldgaber, D. & Housman, D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl Acad. Sci. USA 96, 11404–11409 (1999).
Perez, M. K. et al. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J. Cell Biol. 143, 1457–1470 (1998).
Cha, J. -H. J. et al. Altered brain neurotransmitter receptors in transgenic mice expressing a portion of an abnormal human Huntington disease gene. Proc. Natl Acad. Sci. USA 95, 6480–6485 (1998). The first study to show abnormal transcriptional regulation in HD.
Kouzarides, T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19, 1176–1179 (2000).
Glozak, M. A., Sengupta, N., Zhang, X. & Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 363, 15–23 (2005).
Norton, V., Marvin, K., Yau, P. & Bradbury, E. Nucleosome linking number change controlled by acetylation of histones H3 and H4. J. Biol. Chem. 265, 19848–19852 (1990).
Lee, D. Y., Hayes, J. J., Pruss, D. & Wolffe, A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72, 73–84 (1993).
Hebbes, T. R., Thorne, A. W., Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7, 1395–1402 (1988).
Rundlett, S. E. et al. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA 93, 14503–14508 (1996).
Zhu, W. G., Lakshmanan, R. R., Beal, M. D. & Otterson, G. A. DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Cancer Res. 61, 1327–1333 (2001).
Marks, P. A., Richon, V. M. & Rifkind, R. A. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl Cancer Inst. 92, 1210–1216 (2000).
Glaser, K. B. et al. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol. Cancer Ther. 2, 151–163 (2003).
Mariadason, J. M., Corner, G. A. & Augenlicht, L. H. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 60, 4561–4572 (2000).
Chambers, A. E. et al. Histone acetylation-mediated regulation of genes in leukaemic cells. Eur. J. Cancer 39, 1165–1175 (2003).
Cress, W. D. & Seto, E. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184, 1–16 (2000).
Timmermann, S., Lehrmann, H., Polesskaya, A. & Harel-Bellan, A. Histone acetylation and disease. Cell. Mol. Life. Sci. 58, 728–736 (2001).
Hughes, R. E. Polyglutamine disease: acetyltransferases awry. Curr. Biol. 12, R141–R143 (2002).
Yang, X. -J. & Gregoire, S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 25, 2873–2884 (2005).
de Ruijter, A. J., van Gennip, A. H., Caron, H. N., Kemp, S. & van Kuilenburg, A. B. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749 (2003). A comprehensive review of the different classes of HDAC.
Taunton, J., Hassig, C. A. & Schreiber, S. L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 (1996).
Finnin, M. S. et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401, 188–193 (1999).
Longworth, M. S. & Laimins, L. A. Histone deacetylase 3 localizes to the plasma membrane and is a substrate of Src. Oncogene 25, 4495–4500 (2006).
Ayer, D. E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9, 193–198 (1999).
Carmen, A. A., Rundlett, S. E. & Grunstein, M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 271, 15837–15844 (1996).
Huang, E. Y. et al. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 14, 45–54 (2000).
Kao, H. Y., Downes, M., Ordentlich, P. & Evans, R. M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev 14, 55–66 (2000).
Fischle, W. et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45–57 (2002). An important study showing HDAC3-dependent activity of class II HDACs.
Seigneurin-Berny, D. et al. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21, 8035–8044 (2001).
Zhang, Y. et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 22, 1168–1179 (2003).
Fischer, D. D. et al. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J. Biol. Chem. 277, 6656–6666 (2002).
Guardiola, A. R. & Yao, T. P. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J. Biol. Chem. 277, 3350–3356 (2002).
Kao, H. Y., Lee, C. H., Komarov, A., Han, C. C. & Evans, R. M. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J. Biol. Chem. 277, 187–193 (2002).
Tong, J. J., Liu, J., Bertos, N. R. & Yang, X. J. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 30, 1114–1123 (2002).
Frye, R. A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 (2000).
Kaeberlein, M., McVey, M. & Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 (1999).
Denu, J. M. Linking chromatin function with metabolic networks: Sir2 family of NAD+-dependent deacetylases. Trends Biochem. Sci. 28, 41–48 (2003).
Sauve, A. A. et al. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40, 15456–15463 (2001).
Sauve, A. A. & Schramm, V. L. SIR2: the biochemical mechanism of NAD+-dependent protein deacetylation and ADP-ribosyl enzyme intermediates. Curr. Med. Chem. 11, 807–826 (2004).
Bitterman, K. J., Anderson, R. M., Cohen, H. Y., Latorre-Esteves, M. & Sinclair, D. A. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277, 45099–45107 (2002).
Anekonda, T. S. & Reddy, P. H. Neuronal protection by sirtuins in Alzheimer's disease. J. Neurochem. 96, 305–313 (2006).
Gao, L., Cueto, M. A., Asselbergs, F. & Atadja, P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 277, 25748–25755 (2002).
Bertos, N. R., Wang, A. H. & Yang, X. J. Class II histone deacetylases: structure, function, and regulation. Biochem. Cell Biol. 79, 243–252 (2001).
Sengupta, N. & Seto, E. Regulation of histone deacetylase activities. J. Cell. Biochem. 93, 57–67 (2004).
McKinsey, T. A., Zhang, C. L. & Olson, E. N. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21, 6312–6321 (2001).
Yang, W. M., Tsai, S. C., Wen, Y. D., Fejer, G. & Seto, E. Functional domains of histone deacetylase-3. J. Biol. Chem. 277, 9447–9454 (2002).
Lu, J., McKinsey, T. A., Nicol, R. L. & Olson, E. N. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl Acad. Sci. USA 97, 4070–4075 (2000).
Miska, E. A. et al. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18, 5099–5107 (1999).
Mao, Z., Bonni, A., Xia, F., Nadal-Vicens, M. & Greenberg, M. E. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science 286, 785–790 (1999).
Gaudilliere, B., Shi, Y. & Bonni, A. RNA Interference reveals a requirement for myocyte enhancer factor 2A in activity-dependent neuronal survival. J. Biol. Chem. 277, 46442–46446 (2002).
Miska, E. A. et al. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 29, 3439–3447 (2001).
Kao, H. Y. et al. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276, 47496–47507 (2001).
Grozinger, C. M. & Schreiber, S. L. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA 97, 7835–7840 (2000).
Bolger, T. A. & Yao, T. P. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J. Neurosci. 25, 9544–9553 (2005).
Chawla, S., Vanhoutte, P., Arnold, F. J., Huang, C. L. & Bading, H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85, 151–159 (2003).
Deckel, A. W., Elder, R. & Fuhrer, G. Biphasic developmental changes in Ca2+/calmodulin-dependent proteins in R6/2 Huntington's disease mice. Neuroreport 13, 707–711 (2002).
Hoshino, M. et al. Histone deacetylase activity is retained in primary neurons expressing mutant huntingtin protein. J. Neurochem. 87, 257–267 (2003).
Sakamoto, J., Miura, T., Shimamoto, K. & Horio, Y. Predominant expression of Sir2α, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 556, 281–286 (2004).
Hisahara, S., Chiba, S., Matsumoto, H. & Horio, Y. Transcriptional regulation of neuronal genes and its effect on neural functions: NAD-dependent histone deacetylase SIRT1 (Sir2α). J. Pharmacol. Sci. 98, 200–204 (2005).
Cha, J. -H. J. Transcriptional dysregulation in Huntington's disease. Trends Neurosci. 23, 387–392 (2000).
Luthi-Carter, R. et al. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum. Mol. Genet. 11, 1911–1926 (2002).
Luthi-Carter, R. et al. Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington's disease mouse models reveal context-independent effects. Hum. Mol. Genet. 11, 1927–1937 (2002).
Hodges, A. et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum. Mol. Genet. 15, 965–977 (2006). The above three papers contain important microarray expression profile information about HD and DRPLA.
Steffan, J. S. et al. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl Acad. Sci. USA 97, 6763–6768 (2000). The first paper to implicate aberrant interactions between CBP and HTT in transcriptional repression in HD.
Nucifora, F. C. Jr et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science 291, 2423–2428 (2001). Highlights the implications of CBP in the pathogenic mechanism of polyglutamine repeat disease.
McCampbell, A. et al. CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet. 9, 2197–2202 (2000).
McCampbell, A. & Fischbeck, K. H. Polyglutamine and CBP: fatal attraction? Nature Med. 7, 528–530 (2001).
Dunah, A. W. et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science 296, 2238–2243 (2002).
van Roon-Mom, W. M. et al. Insoluble TATA-binding protein accumulation in Huntington's disease cortex. Brain Res. Mol. Brain Res. 109, 1–10 (2002).
Igarashi, S. et al. Inducible PC12 cell model of Huntington's disease shows toxicity and decreased histone acetylation. Neuroreport 14, 565–568 (2003).
Hazeki, N. et al. Ultrastructure of nuclear aggregates formed by expressing an expanded polyglutamine. Biochem. Biophys. Res. Commun. 294, 429–440 (2002).
Mori, N., Schoenherr, C., Vandenbergh, D. J. & Anderson, D. J. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron 9, 45–54 (1992).
Schoenherr, C. J., Paquette, A. J. & Anderson, D. J. Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl Acad. Sci. USA 93, 9881–9886 (1996).
Chen, Z. -F., Paquette, A. J. & Anderson, D. J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nature Genet. 20, 136–142 (1998).
Zuccato, C. et al. Loss of huntingtin-mediated BDNF gene transcription in huntington's disease. Science 293, 493–498 (2001).
Zuccato, C. et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. 35, 76–83 (2003).
Bates, E. A., Victor, M., Jones, A. K., Shi, Y. & Hart, A. C. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J. Neurosci. 26, 2830–2838 (2006).
Hubbert, C. et al. HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 (2002).
Bali, P. et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J. Biol. Chem. 280, 26729–26734 (2005).
Kovacs, J. J. et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell 18, 601–607 (2005).
Hook, S. S., Orian, A., Cowley, S. M. & Eisenman, R. N. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl Acad. Sci. USA 99, 13425–13430 (2002).
Hideshima, T. et al. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc. Natl Acad. Sci. USA 102, 8567–8572 (2005).
Iwata, A., Riley, B. E., Johnston, J. A. & Kopito, R. R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280, 40282–40292 (2005).
Kawaguchi, Y. et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738 (2003).
Grubisha, O., Smith, B. C. & Denu, J. M. Small molecule regulation of Sir2 protein deacetylases. FEBS J. 272, 4607–4616 (2005).
Ghosh, S. & Feany, M. B. Comparison of pathways controlling toxicity in the eye and brain in Drosophila models of human neurodegenerative diseases. Hum. Mol. Genet. 13, 2011–2018 (2004).
Parker, J. A. et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nature Genet. 37, 349–350 (2005).
Sinclair, D. Sirtuins for healthy neurons. Nature Genet. 37, 339–340 (2005).
Levine, A. J. p53, the cellular gatekeeper for growth and division. Cell 88, 323–331 (1997).
Hughes, P. E., Alexi, T. & Schreiber, S. S. A role for the tumour suppressor gene p53 in regulating neuronal apoptosis. Neuroreport 8, v–xii (1997).
Gu, W. & Roeder, R. G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 (1997).
Ito, A. et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21, 6236–6245 (2002).
Gang Liu, X. C. Regulation of the p53 transcriptional activity. J. Cell. Biochem. 97, 448–458 (2006).
Luo, J. et al. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl Acad. Sci. USA 101, 2259–2264 (2004).
Bae, B. I. et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron 47, 29–41 (2005).
Feng, D. & Kan, Y. W. The binding of the ubiquitous transcription factor Sp1 at the locus control region represses the expression of β-like globin genes. Proc. Natl Acad. Sci. USA 102, 9896–9900 (2005).
Murphy, M. et al. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13, 2490–2501 (1999).
Luo, J., Su, F., Chen, D., Shiloh, A. & Gu, W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408, 377–381 (2000).
Juan, L. J. et al. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275, 20436–20443 (2000).
Pratt, W. B. & Toft, D. O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228, 111–133 (2003).
Bagatell, R. et al. Destabilization of steroid receptors by heat shock protein 90-binding drugs: a ligand-independent approach to hormonal therapy of breast cancer. Clin. Cancer Res. 7, 2076–2084 (2001).
Neckers, L. Heat shock protein 90 inhibition by 17-allylamino-17- demethoxygeldanamycin: a novel therapeutic approach for treating hormone-refractory prostate cancer. Clin. Cancer Res. 8, 962–966 (2002).
Waza, M. et al. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nature Med. 11, 1088–1095 (2005).
Aoyagi, S. & Archer, T. K. Modulating molecular chaperone Hsp90 functions through reversible acetylation. Trends Cell Biol. 15, 565–567 (2005).
Chen, L. et al. Chemical ablation of androgen receptor in prostate cancer cells by the histone deacetylase inhibitor LAQ824. Mol. Cancer Ther. 4, 1311–1319 (2005).
Ren, M., Leng, Y., Jeong, M., Leeds, P. R. & Chuang, D. M. Valproic acid reduces brain damage induced by transient focal cerebral ischemia in rats: potential roles of histone deacetylase inhibition and heat shock protein induction. J. Neurochem. 89, 1358–1367 (2004).
Zhao, Y. et al. Lifespan extension and elevated hsp gene expression in Drosophila caused by histone deacetylase inhibitors. J. Exp. Biol. 208, 697–705 (2005).
Hay, D. G. et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 13, 1389–1405 (2004).
Woodman, B. et al. The HdhQ150/Q150 knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res. Bull. (in the press).
Li, F., Macfarlan, T., Pittman, R. N. & Chakravarti, D. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J. Biol. Chem. 277, 45004–45012 (2002).
Gatchel, J. R. & Zoghbi, H. Y. Diseases of unstable repeat expansion: mechanisms and common principles. Nature Rev. Genet. 6, 743–755 (2005).
Tsai, C. C. et al. Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc. Natl Acad. Sci. USA 101, 4047–4052 (2004).
Matilla, A. et al. The cerebellar leucine-rich acidic nuclear protein interacts with ataxin-1. Nature 389, 974–978 (1997).
Seo, S. et al. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104, 119–130 (2001).
Chen, H. K. et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell 113, 457–468 (2003).
Burnett, B., Li, F. & Pittman, R. N. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 12, 3195–3205 (2003).
Burnett, B. G. & Pittman, R. N. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc. Natl Acad. Sci. USA 102, 4330–4335 (2005).
Palhan, V. B. et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl Acad. Sci. USA 102, 8472–8477 (2005).
McMahon, S. J., Pray-Grant, M. G., Schieltz, D., Yates, J. R. & Grant, P. A. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc. Natl Acad. Sci. USA 102, 8478–8482 (2005).
Sterner, D. E., Belotserkovskaya, R. & Berger, S. L. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl Acad. Sci. USA 99, 11622–11627 (2002).
Pray-Grant, M. G. et al. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22, 8774–8786 (2002).
Helmlinger, D. et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 4, e67 (2006).
Schilling, G. et al. Nuclear accumulation of truncated atrophin-1 fragments in a transgenic mouse model of DRPLA. Neuron 24, 275–286 (1999).
Wood, J. D. et al. Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription. J. Cell Biol. 150, 939–948 (2000).
Gelmetti, V. et al. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 18, 7185–7191 (1998).
Lutterbach, B. et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 18, 7176–7184 (1998).
Wang, J., Hoshino, T., Redner, R. L., Kajigaya, S. & Liu, J. M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl Acad. Sci. USA 95, 10860–10865 (1998).
Ying, M. et al. Sodium butyrate ameliorates histone hypoacetylation and neurodegenerative phenotypes in a mouse model for DRPLA. J. Biol. Chem. 281, 12580–12586 (2006).
Wang, L., Rajan, H., Pitman, J. L., McKeown, M. & Tsai, C. -C. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 20, 525–530 (2006).
McCampbell, A. et al. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc. Natl Acad. Sci. USA 98, 15179–15184 (2001).
Steffan, J. S. et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413, 739–743 (2001). The first study in Drosophila to show HDAC inhibitors as a beneficial therapy for polyglutamine repeat disease.
Hockly, E. et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc. Natl Acad. Sci. USA 100, 2041–2046 (2003). The first published study of the use of an HDAC inhibitor as a therapeutic agent in a mouse model of HD.
Ferrante, R. J. et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J. Neurosci. 23, 9418–9427 (2003).
Gardian, G. et al. Neuroprotective effects of phenylbutyrate in the N171–82Q transgenic mouse model of Huntington's disease. J. Biol. Chem. 280, 556–563 (2005).
Drummond, D. C. et al. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu. Rev. Pharmacol. Toxicol. 45, 495–528 (2005).
Miller, T. A., Witter, D. J. & Belvedere, S. Histone deacetylase inhibitors. J. Med. Chem. 46, 5097–5116 (2003).
Grozinger, C. M. & Schreiber, S. L. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 9, 3–16 (2002).
Vannini, A. et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl Acad. Sci. USA 101, 15064–15069 (2004).
Minamiyama, M. et al. Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 13, 1183–1192 (2004).
Borovecki, F. et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc. Natl Acad. Sci. USA 102, 11023–11028 (2005).
Agrawal, N. et al. Identification of combinatorial drug regimens for treatment of Huntington's disease using Drosophila. Proc. Natl Acad. Sci. USA 102, 3777–3781 (2005).
Byers, R. K. & Banker, B. Q. Infantile muscular atrophy. Arch. Neurol. 5, 140–164 (1961).
Brzustowicz, L. M. et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2–13.3. Nature 344, 540–541 (1990).
Melki, J. et al. Gene for chronic proximal spinal muscular atrophies maps to chromosome 5q. Nature 344, 767–768 (1990).
Gilliam, T. C. et al. Genetic homogeneity between acute and chronic forms of spinal muscular atrophy. Nature 345, 823–825 (1990).
Lefebvre, S. et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nature Genet. 16, 265–269 (1997).
Iannaccone, S. T., Smith, S. A. & Simard, L. R. Spinal muscular atrophy. Curr. Neurol. Neurosci. Rep. 4, 74–80 (2004).
Chang, J. G. et al. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl Acad. Sci. USA 98, 9808–9813 (2001).
Sumner, C. J. et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 54, 647–654 (2003).
Brichta, L. et al. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 12, 2481–2489 (2003).
Andreassi, C. et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur. J. Hum. Genet. 12, 59–65 (2004).
Riessland, M. Brichta, L., Hahnen, E. & Wirth, B. The benzamide M344, a novel histone deacetylase inhibitor, significantly increases SMN2 RNA/protein levels in spinal muscular atrophy cells. Hum. Genet. 1–10 (2006).
Kernochan, L. E. et al. The role of histone acetylation in SMN gene expression. Hum. Mol. Genet. 14, 1171–1182 (2005).
Jung, M. et al. Amide analogues of trichostatin A as inhibitors of histone deacetylase and inducers of terminal cell differentiation. J. Med. Chem. 42, 4669–4679 (1999).
Furumai, R. et al. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc. Natl Acad. Sci. USA 98, 87–92 (2001).
Roopra, A. et al. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell. Biol. 20, 2147–2157 (2000).
Kao, H. Y. et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12, 2269–2277 (1998).
Zhang, Y. et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13, 1924–1935 (1999).
Amann, J. M. et al. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21, 6470–6483 (2001).
Wen, Y. D. et al. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl Acad. Sci. USA 97, 7202–7207 (2000).
Li, J. et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19, 4342–4350 (2000).
Grozinger, C. M., Hassig, C. A. & Schreiber, S. L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. PNAS 96, 4868–4873 (1999).
Waltregny, D. et al. Histone deacetylase HDAC8 associates with smooth muscle alpha-actin and is essential for smooth muscle cell contractility. FASEB J. 19, 966–998 (2005).
Lemercier, C. et al. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275, 15594–15599 (2000).
Huntington's Disease Collaborative Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 (1993).
Spada, A. R. L., Wilson, E. M., Lubahn, D. B., Harding, A. E. & Fischbeck, K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352, 77–79 (1991).
Koide, R. et al. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA). Nature Genet. 6, 9–13 (1994).
Nagafuchi, S. et al. Dentatorubral and pallidoluysian atrophy expansion of an unstable CAG trinucleotide on chromosome 12p. Nature Genet. 6, 14–18 (1994).
Orr, H. T. et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nature Genet. 4, 221–226 (1993).
Matilla, T. et al. Presymptomatic analysis of spinocerebellar ataxia type 1 (SCA1) via the expansion of the SCA1 CAG-repeat in a large pedigree displaying anticipation and parental male bias. Hum. Mol. Genet. 2, 2123–2128 (1993).
Imbert, G. et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nature Genet. 14, 285–291 (1996).
Sanpei, K. et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nature Genet. 14, 277–284 (1996).
Pulst, S. -M. et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nature Genet. 14, 269–276 (1996).
Kawaguchi, Y. et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. 8, Nature Genet. 221–228 (1994).
Zhuchenko, O. et al. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the α1A-voltage-dependent calcium channel. Nature Genet. 15, 62–69 (1997).
David, G. et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nature Genet. 17, 65–70 (1997).
Lindblad, K. et al. An expanded CAG repeat sequence in spinocerebellar ataxia type 7. Genome Res. 6, 965–971 (1996).
Koide, R. et al. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum. Mol. Genet. 8, 2047–2053 (1999).
Schaffar, G. et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell 15, 95–105 (2004).
Acknowledgements
Work in the author's laboratory is funded by the Wellcome Trust, Hereditary Disease Foundation, Huntington's Disease Society of America Coalition for the Cure and the HighQ Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Histidine-aspartate charge-relay system
-
Histidine and aspartate residues coordinate the zinc cation and water molecule within the active site of the histone deacetylase enzyme that is required for the cleavage of an acetyl group from the substrate.
- Nuclear co-repressor/silencing mediator of retinoid and thyroid hormone receptors
-
(N-CoR/SMRT). Co-repressors that interact with unliganded nuclear receptors and form repressor complexes that recruit histone deacetylases and other proteins to actively repress transcription.
- Repressor element 1 transcription factor/neuron restrictive silencer factor
-
(REST/NSRF). REST/NRSF is a negative regulator of neuronal genes that contains a specific DNA response element in the promoter, the neuron restrictive silencing element (NRSE). REST/NRSF ensures the correct patterning of neuronal gene expression and the silencing of these genes in non-neuronal tissues.
- Neuron restrictive silencing element
-
(NRSE). An NRSE is a 21 bp silencer element found in the promoters of neuronal genes to which REST/NRSF binds. It is also known as repressor element 1 (RE1).
- RNA interference
-
(RNAi). A method by which double-stranded RNA that is encoded on an exogenous vector can be used to interfere with normal RNA processing, causing rapid degradation of the endogenous RNA and thereby precluding translation. This provides a simple way of studying the effects of the absence of a gene product in simple organisms and cells.
- Dynein
-
A microtubule minus-end-directed motor protein that carries out retrograde transport from the cell surface to the centre of the cell. Dynein is a large molecule that complexes with dynactin to form direct and indirect associations with various cargoes.
- Aggresome
-
An inclusion body of aggregated material, usually perinuclear, formed through dynein-dependent retrograde transport on microtubules.
- Co-chaperones
-
Proteins associated with molecular chaperones that are necessary for full functional activity of the chaperone.
- Small-molecule hydroxamates
-
Inhibitors containing hydroxamic acid residues are potent but reversible inhibitors of class I/II histone deacetylases (HDACs). They bind with high affinity and chelate the zinc cation at the bottom of the HDAC catalytic pocket.
- Cyclic peptides
-
Histone deacetylase inhibitors that have epoxyketone (epoxide) moieties that could act by chemically binding to either the zinc cation or an amino acid in the active site.
- Benzamides
-
A structurally diverse group of compounds that have an unknown mechanism of action. The diaminophenyl group present in these compounds is thought to be important for their inhibitory behaviour.
Rights and permissions
About this article
Cite this article
Butler, R., Bates, G. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat Rev Neurosci 7, 784–796 (2006). https://doi.org/10.1038/nrn1989
Issue Date:
DOI: https://doi.org/10.1038/nrn1989
This article is cited by
-
Exploring the combined anti-cancer effects of sodium butyrate and celastrol in glioblastoma cell lines: a novel therapeutic approach
Medical Oncology (2024)
-
Injured adult neurons regress to an embryonic transcriptional growth state
Nature (2020)
-
HDAC1 inhibition ameliorates TDP-43-induced cell death in vitro and in vivo
Cell Death & Disease (2020)
-
Molecular Targets and Therapeutic Strategies in Spinocerebellar Ataxia Type 7
Neurotherapeutics (2019)
-
The histone deacetylase inhibitor, sodium butyrate, exhibits neuroprotective effects for ischemic stroke in middle-aged female rats
Journal of Neuroinflammation (2016)