Abstract
Almost all human proteins are subject to proteolytic degradation, which produces a broad range of peptides that have highly specific and sometimes unexpected functions. Peptide libraries that have been generated from human bodily fluids or tissues are a rich but mostly unexplored source of bioactive compounds that could be used to develop antimicrobial and immunomodulatory therapeutic agents. In this Innovation article, we describe the discovery, optimization and application of endogenous bioactive peptides from human-derived peptide libraries, with a particular focus on the isolation of endogenous inhibitors and promoters of HIV-1 infection.
Similar content being viewed by others
Main
The human genome contains only about 21,000 distinct protein-coding genes1, which is substantially fewer than originally expected. However, owing to alternative splicing mechanisms and many post-translational modifications, these ∼21,000 genes are estimated to produce more than one million different proteins2. Although the proteome (that is, the whole set of proteins that are expressed in a specified cell, tissue or organism) is highly complex, it is outnumbered by the many smaller peptides that are generated from full-length proteins by proteolytic cleavage. The largest fraction of the peptidome (that is, all of the peptides that are present in a specific cell, tissue, organism or system) comprises peptide fragments that are generated by the proteolysis of larger precursor molecules. Only a small fraction of the peptidome has been functionally characterized, owing to its enormous complexity and the lack of broadly applicable methods that enable its unbiased analysis.
For many years, the peptidome was commonly regarded as 'biological trash'. However, it has recently been recognized that the expression and activities of the more than 500 human proteases that generate these peptides are altered under pathological conditions3. Furthermore, accumulating evidence suggests that some cleavage products of larger precursor proteins exert specific and sometimes highly unexpected activities against human pathogens4,5,6,7,8,9,10,11,12,13,14. Thus, the human peptidome is a rich source of disease-specific biomarkers. It has long been known that humans and other mammalian species have evolved various antimicrobial peptides as a first line of defence against viruses, bacteria and fungi15,16,17,18. Although the role of some antimicrobial peptides, such as defensins and cathelicidins, in innate immunity, immune modulation and inflammation is well-established15,16,17,18, it seems highly likely that many important peptidic immune modulators and effectors in the human body remain to be identified.
Thousands of antimicrobial peptides have been isolated from various natural sources (for example, bacteria, fungi, plants and animals). Furthermore, several inhibitors of bacterial infections have been isolated from human immune cells (for example, α-defensins)19, skin (for example, β-defensins)20, saliva (for example, histatins)21, nasal secretions (for example, lysozyme)22, seminal plasma (for example, cathelicidins)23,24 and sweat (for example, dermicidin)25. The various types of antimicrobial peptides15,16,17,18,19,20,21,22,23,24,25 and their possible clinical applications26,27,28,29,30,31 have been the topics of several recent reviews. In this Innovation article, we discuss the discovery of novel antimicrobial peptides by the systematic screening of highly complex peptide libraries that are derived from human bodily fluids or tissues. In particular, we focus on the isolation of endogenous agents that affect HIV-1 infection by unexpected mechanisms and that may have implications beyond HIV/AIDS. Furthermore, we present the advantages and disadvantages of this strategy. Finally, we discuss how endogenous peptides that have been isolated from the human peptidome can be optimized for therapeutic or basic research applications, and we highlight future directions and challenges.
Human-derived peptide libraries
Peptide libraries that have been generated from human bodily fluids or tissues, such as blood or lymphoid tissues, contain essentially all peptides and small proteins of the respective source in their final processed and physiologically relevant forms32,33,34. Thus, they are an excellent source for the identification of as-yet-unknown bioactive peptides.
Bioactive peptides from human sources. Most clinically approved drugs are small molecules, and most of these compounds have a molecular weight of much less than 1 kDa. Such a small size is one of the requirements for uptake through the gastrointestinal tract and thus oral application. By contrast, large protein drugs, such as antibodies, have molecular weights of up to 150 kDa and need to be administered parenterally. The size of peptides, which are usually up to 50 amino acids in length, is between these two drug classes, although there is no strict demarcation28,29,30,31. Antimicrobial molecules are found in all three size classes, including small molecules such as penicillin (which is 0.334 kDa), the glycopeptide vancomycin (which is 1.449 kDa) and large rabies-specific immunoglobulins (which are about 150–170 kDa)28. Other examples of clinically approved antimicrobial peptides are the antibiotics polymyxin B and polymyxin E (also known as colistin) and the antiviral agent interferon-α. Owing to the increasing resistance of microorganisms to conventional antibiotics, there is currently a growing interest in the development of antimicrobial peptides for clinical applications, and several candidates are in clinical trials or under development26,27,28,29,30,31. The Antimicrobial Peptide Database (see Further information) provides an overview of bioactive antibacterial and antiviral peptides that have been isolated from natural sources. Peptide drugs may be less toxic and are less likely to cause allergic or inactivating immune responses than small-molecule or protein drugs, particularly if they are derived from a human source16. However, the isolation of individual bioactive peptides from highly complex human bodily fluids or tissues is a challenging task.
Generating peptide libraries from human haemofiltrate. Human tissues and bodily fluids are usually only available in very limited quantities, and standardized methods to purify bioactive agents from the large number of highly diverse peptides and proteins that are contained in these tissues and fluids are mostly unavailable. Large quantities of starting materials are advantageous from a technical and experimental standpoint. One abundant source of human peptides is haemofiltrate, which is a waste product of dialysis that contains essentially all blood components that have a molecular weight of less than 20–30 kDa (Box 1) and is available from patients with chronic renal failure at quantities of thousands of litres. A combination of ultrafiltration, followed by cation-exchange separation and reverse-phase chromatography, enables the standardized separation and concentration of all of the peptides and small proteins that are present in haemofiltrate into about 300–500 fractions32,33 (Box 1). The analysis of these fractions does not require the high-throughput methodologies and facilities that are required for the testing of other libraries, which often comprise hundreds of thousands of compounds. For example, specific assays that test the inhibition of a particular pathogen, the modulation of specific cellular functions or the induction of selected immune factors can be used to identify and purify the most active agents that are present in these fractions. Such peptide libraries are a useful source for the discovery of novel bioactive agents, as they represent the enormous structural and functional diversity of the human peptidome and the peptides are present in their final processed, and thus bioactive, forms32,33,34,35. Notably, haemofiltrate contains not only endocrine peptides but also peptides that function in a paracrine or autocrine manner, as a small fraction of these peptides are released into the extracellular space and are found in the blood34,35. Thus, haemofiltrate-derived peptide libraries contain essentially the entire circulating blood peptidome in a lyophilized, bioactive and highly concentrated form. However, in addition to peptides that naturally exist in the human body, these libraries may also contain proteolytic cleavage products that specifically arise and accumulate during the collection or storage of body fluids or tissues. Nonetheless, these peptide libraries are an excellent source for the identification of endogenous bioactive peptides.
Alternative human sources. Large quantities of starting material are advantageous but not always obligatory for the identification of novel endogenous bioactive peptides. In fact, many peptides or proteins only become active and exert their respective functions in specific compartments or at sites of infection and/or inflammation; for example, saliva, genital fluid, milk and sweat contain particularly large numbers of antimicrobial peptides16. To isolate agents that are not circulating in the bloodstream, it is important to generate peptide libraries from sources other than haemofiltrate, and in principle, they can be produced from any tissue or bodily fluid. Using ultrafiltration and cation exchange followed by reverse-phase chromatography, peptide libraries have been generated from human plasma, milk, placenta, sweat, seminal plasma and saliva (J.M., L.S., W.-G.F. and F.K., unpublished observations), and some antimicrobial peptides that have been isolated from these libraries are listed in Table 1.
Human peptides that affect HIV-1 infection
Most clinically used antiretroviral drugs are small molecules that have been selected to specifically target and inhibit viral proteins that are essential for HIV-1 replication. Enfuvirtide (also known as T-20; Trimeris) is the only currently licensed peptidic antiretroviral drug36. Enfuvirtide is derived from an α-helical region in the viral transmembrane glycoprotein known as heptad repeat 2 (HR2). By mimicking HR2 and competitively binding to another α-helical region of gp41, known as HR1, enfuvirtide prevents the interaction between HR1 and HR2 and thus prevents the conformational change that is required for virion fusion with the host cell37. Although this protein is not from a human source, enfuvirtide provides an example of the feasibility of developing peptide antiretroviral agents. In addition, chemokines, such as CXC-chemokine ligand 12 (CXCL12; also known as SDF-1) and CC-chemokine ligand 3 (CCL3; also known as MIP-1α), CCL4 (also known as MIP-1β) or CCL5 (also known as RANTES), bind to the G protein-coupled receptors (GPCRs) CXC-chemokine receptor 4 (CXCR4) and CC-chemokine receptor 5 (CCR5), respectively, and they prevent infection by HIV-1 strains that use the same receptors for viral entry. In fact, these inhibitory activities were crucial for the discovery of CCR5 as the main co-receptor for HIV-1 entry38,39. Modified derivatives of CCL5 are extremely potent inhibitors of R5-tropic HIV-1 strains in vitro40, but they have a low efficiency in vivo. Several intracellular host proteins that inhibit HIV-1 replication at different steps of the viral life cycle41 — which are known as restriction factors — have more recently been discovered, and it seems highly likely that additional antiretroviral peptides and/or proteins remain to be discovered.
Identification of novel endogenous HIV-1 inhibitors. An initial screen of a haemofiltrate-derived peptide library for CCR5 ligands identified a naturally occurring truncated form of the abundant human chemokine CCL14 as a potent agonist of the receptor, which inhibits infection by R5-tropic HIV-1 strains6,42. Notably, other chemokines, such as CXCL4 and specific fragments of CXCL4, also inhibit HIV-1 (Ref. 43) and other important pathogens, such as Plasmodium falciparum44. Screening of further haemofiltrate-derived peptide fractions identified another HIV-1 inhibitor: a 20-residue C-proximal subfragment of the serine protease α1-antitrypsin, which has been designated virus-inhibitory peptide (VIRIP). This peptide was purified by multiple rounds of peptide separation and antiviral screening8 (Fig. 1). VIRIP specifically interacts with the viral fusion peptide that is located at the amino terminus of the gp41 transmembrane domain, which is a region that is distinct from the HR1 region that is targeted by enfuvirtide. VIRIP blocks an entry step before membrane fusion — that is, penetration and insertion of the viral fusion peptide into the host cell membrane (Fig. 2). This is a novel and unexpected mechanism, as the fusion peptide only becomes accessible during the viral entry process after conformational changes of gp41 (Ref. 45). Thus, it was assumed that this anchoring step cannot be inhibited45. The finding that viral fusion peptides can be targeted by an antiviral agent may offer perspectives beyond HIV/AIDS, as many enveloped viruses, such as influenza virus, hepatitis C virus, filovirus, coronavirus, arenavirus and hepadnaviruses use fusion peptides to infect their target cells46,47,48,49,50. Owing to this distinct inhibitory mechanism, VIRIP is active against HIV-1 strains that are resistant to other antiretroviral drugs, including other entry inhibitors8. A proteomics approach identified VIRIP in plasma from patients with acute HIV-1 infection, which suggests that it may contribute to the earliest systemic antiviral response in HIV-1 infection51. Thus, screening of haemofiltrate-derived peptide libraries enabled the isolation of two novel HIV-1 inhibitors: a CCR5 agonist and a peptide that targets the HIV-1 gp41 fusion peptide (Fig. 2). It is remarkable that this approach did not identify the well-known CCR5 ligand CCL5 but instead identified a novel agonist, and this suggests that other relevant peptide ligands of GPCRs that are targets of many modern drugs remain to be discovered.
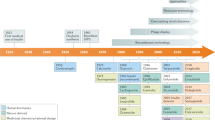
Schematic outline of the general strategy to isolate unknown antiviral peptides from human peptide libraries. The isolation of the HIV-1 inhibitor virus-inhibitory peptide (VIRIP)8 is shown as an example. pH pools that have been obtained using cation exchange are subjected to reverse-phase chromatography, and the resulting fractions are assayed for antiviral activity (for example, effects on HIV-1 replication). The active fraction (indicated in red) is further purified by multiple rounds of chromatography. Each cycle of separation and screening reduces the complexity of the peptide samples until the purified bioactive agent can be identified by mass spectrometry analysis and peptide sequencing. VIRIP was shown to be a 20-residue (LEAIPMSIPPEVKFNKPFVF) fragment of the serine protease α1-antitrypsin8. The identified peptides can be functionally characterized and optimized for clinical development.
Overview of the HIV-1 entry steps that are targeted by peptides. Amyloidogenic peptides that are derived from human semen, such as fragments of prostatic acidic phosphatase (PAP), can self-assemble into amyloid fibrils that promote the attachment of the virus to target cells by neutralizing the repulsion between negatively charged viral and cellular membranes8,61. Unlike these semen-derived enhancing agents, human peptides that are isolated from haemofiltrate have antiviral activity. The truncated form of the abundant human chemokine CC-chemokine ligand 14 (CCL14(9–74)) inhibits entry of R5-tropic HIV-1 strains by binding to the CC-chemokine receptor 5 (CCR5) co-receptor6,42. Human-derived virus-inhibitory peptide (VIRIP; which is a subfragment of the serine protease inhibitor α1-antitrypsin) also targets the fusion peptide by specifically interacting with the amino terminus of its transmembrane domain to block penetration and insertion of the viral fusion peptide8. Enfuvirtide is a synthetic peptide that is derived from an α-helical region in the HIV-1 transmembrane glycoprotein36. This inhibitor binds to helical repeat region 1 (HR1) of gp41, which is a region that is distinct from the region that is targeted by VIRIP, and prevents the formation of a hairpin structure that is required for membrane fusion, thereby blocking HIV-1 infection. The target sites for VIRIP and enfuvirtide in gp41 are indicated. From Forssman, W.-G. et al. Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide. Sci. Transl. Med. 2, 63re3 (2010). Reprinted with permission from AAAS.
Endogenous peptides that promote HIV-1 infection. The success of screening haemofiltrate-derived peptide libraries encouraged the screening of smaller-scale libraries from other sources for novel antiviral agents. The efficiency of virus transmission by sexual contact is surprisingly low, which suggests that inhibitors of HIV-1 infection are present in semen. Furthermore, it has been reported that semen contains various antimicrobial agents23,24,52,53,54,55 and cationic peptides that have anti-HIV-1 activity56. However, unexpectedly, screening of a semen-derived peptide library only identified fractions that promoted HIV-1 infection57, which might reflect the specific adaptation of the virus for efficient transmission in this environment. Further analyses showed that C-proximal fragments of prostatic acidic phosphatase (PAP), which is an abundant enzyme in semen, self-assemble into cationic amyloid fibrils57. These fibrils, which are known as semen-derived enhancers of virus infection (SEVIs), increase the infectious virus titre in cell culture by several orders of magnitude57 and can facilitate virus infection in non-human primate models after exposure to low viral doses that approximate the in vivo situation58. Virus attachment is usually ineffective as the densities of the viral envelope glycoproteins on the virion and the CD4 receptor on the target cell are low and the surfaces of viral and cellular membranes are negatively charged and usually repel each other. Both SEVI fibril formation and its positive surface charge are crucial for increased HIV-1 infection59, which suggests that SEVI promotes the attachment of the virus to target cells by neutralizing the repulsion between negatively charged viral and cellular membranes (Fig. 2). Interestingly, follow-up studies showed that an N-proximal fragment of PAP, as well as peptides that are derived from semenogelin 1 and semenogelin 2, which are the most abundant proteins in semen, also form amyloid fibrils that increase HIV-1 infection60,61. Endogenous amyloid fibrils have recently been detected and structurally characterized in fresh non-modified ejaculates62. Notably, their levels correlate with the efficiency of semen-mediated promotion of HIV-1 infection in vitro61,63. Thus, semen, which is the main vector for HIV-1 transmission, contains amyloid fibrils that boost viral infectivity and are therefore novel targets to reduce the risk of sexual transmission. In fact, several agents that block the formation or infection-promoting activity of semen-derived fibrils have been reported64,65,66,67,68. Such amyloids may also be exploited by other pathogens, as recent data show that semen amyloids also increase cytomegalovirus (CMV) infection69. Semen is the first bodily fluid that has been identified to naturally contain amyloid fibrils in a non-diseased state62. The physiological role of seminal amyloids in healthy individuals remains to be defined; however, it is tempting to speculate that they might be involved in both innate immunity70 and fertility.
Optimization and development. VIRIP and CCL14(9–74) are both active against HIV-1 in the lower micromolar range. This concentration may well be physiologically relevant, as it can readily be achieved by proteolytic cleavage of their highly abundant α1-antitrypsin and CCL14 precursors. However, this concentration is suboptimal for therapeutic applications, which usually require activities in the nanomolar range71. Specific modifications of these natural HIV-1 inhibitors increased their antiviral activity by about two orders of magnitude. Chemical modifications of the N terminus of CCL14(9–74) rendered the peptide resistant to degradation and thereby strongly increased its antiretroviral potency72. In the case of VIRIP, the introduction of amino acid alterations that stabilize the active conformation of the peptide and increase its hydrophobic interactions with the HIV-1 fusion peptide greatly increased its antiviral potency8. Notably, short-term monotherapy with one of these optimized derivatives (known as VIR-576) was well tolerated and reduced plasma viral loads in treatment-naive patients with HIV-1 (Ref. 73). This provides proof-of-concept that naturally occurring antiviral peptides can be optimized for therapeutic applications. Owing to their mode of application (that is, injection) and the high costs that are involved in peptide synthesis, VIRIP derivatives are not yet competitive with the many highly effective and orally available small-molecule drugs that are available for the treatment of HIV/AIDS. Several strategies are underway to further increase the efficacy and application of VIRIP derivatives, including packaging them into nanoparticles to improve their delivery and stability, as well as structure-based optimization.
Semen-derived amyloid fibrils have also been developed for further applications; as they boost HIV-1 infection, they could be used to improve retroviral gene delivery. Indeed, SEVI increases the efficiency of retroviral gene transfer, although only with moderate and variable efficiency74. On the basis of these findings, broader screens were carried out for amyloidogenic peptides that function as promoters of retroviral transduction. These studies led to the development of a synthetic 12-residue peptide known as EF-C (Protransduzin; Pharis Biotec GmbH) that instantaneously self-assembles into nanofibrils and promotes retroviral gene transfer more efficiently than semen-derived fibrils75. Furthermore, EF-C captures virions and thus enables retroviral particles to be concentrated by conventional low-speed centrifugation instead of the more laborious ultracentrifugation, which has so far been used for virus concentration. These developments pave the way for a new class of promoters of retroviral gene transfer in basic research and clinical applications.
Proteolytic cleavage as a common defence mechanism? It is remarkable that a single assay system (that is, the inhibition of HIV-1 infection) led to the discovery of three novel endogenous agents that affect HIV-1 entry by distinct mechanisms (Fig. 2). As discussed above, these peptides are structurally and functionally distinct from other positively charged amphipathic antimicrobial peptides and might have implications in addition to HIV/AIDS in innate and adaptive immunity as well as in sperm function and fertility. It is also striking that these newly identified modulators of HIV-1 infection are generated from highly abundant precursor proteins: α1-antitrypsin, PAP and semenogelins. The proteases that generate the bioactive peptides remain to be fully defined. However, at least in some cases, these proteases are released from immune cells and are induced or activated by inflammation; for example, VIRIP is generated by matrix metalloproteinase 9 (MMP9)8 and CCL14(9–74) is generated by serine proteases6. Furthermore, these endogenous peptides interact with their respective targets in a highly specific manner, and subtle structural changes at the N terminus (for example, removing or adding single amino acid residues) disrupt their functional activities6,8. Thus, it is tempting to speculate that the cleavage of abundant precursors may be a more common, fast and effective means to generate immunomodulators and antimicrobial peptides at the locations and times that they are actually needed, and it is likely that such peptides have roles in various biological processes. Moreover, the rapid generation of specific effectors by the proteolysis of abundant precursor proteins instead of de novo protein synthesis may be particularly advantageous for innate defence mechanisms against invading pathogens for which immediate responses are crucial for effective counteraction. Notably, the local extracellular acidification that activates various proteases is currently emerging as a key regulatory concept of innate immunity76,77.
Other antimicrobial peptides. The use of peptide libraries from human sources for the discovery of novel factors that modulate HIV-1 infection is a prime example of peptide drug discovery; however, in principle, such libraries can be used to identify endogenous peptides that have any activity of interest. The main prerequisite is a robust bioassay that enables the reliable screening of several hundreds of peptide fractions. A list of antiviral and antibacterial peptides that have been identified by screening human peptide libraries is provided in Table 1. These agents were isolated from various sources, such as haemofiltrate, placental tissue, milk and semen, and target various bacterial and viral pathogens. This list also includes human β-defensin 1 (Ref. 78) — the first member of the family of β-defensins to be discovered in humans — which mediates the resistance of epithelial surfaces to microbial colonization. Several agents that have been isolated from human-derived peptide libraries show the typical features of most antimicrobial peptides — that is, they are cationic and amphipathic, and destabilize the membranes of pathogens (Table 1). The microbicidal action of other endogenous peptides that were identified in this screen was less predictable; for example, a recently discovered derivative of the neutrophil-activating peptide 2 (known as CYVIP) inhibits human CMV (HCMV) infection by binding to heparan sulphate proteoglycans, which function as attachment receptors for CMV79. Moreover, an endogenous human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) peptide has been isolated from human placental tissue. This fragment is internalized by Candida albicans and kills the pathogen by inducing rapid apoptotic death13. As these discoveries are based on only a limited number of antimicrobial screens, this suggests that the human peptidome might yield more surprising findings.
Obstacles to purifying bioactive peptides. Currently, a key challenge is the purification and identification of the active compounds from the complex peptide mixture in active fractions of the generated peptide libraries. However, rapid progress has been made in the development of improved methodologies for peptide separation and analysis80; for example, state-of-the-art mass spectrometry methods enable the convenient deduction of peptide sequences from complex fragmentation spectra. Furthermore, highly sensitive time-of-flight (TOF) mass spectrometers for matrix-assisted laser desorption–ionization (MALDI) and electrospray ionization (ESI) already enable accurate mass determination of peptides at femtomolar concentrations. The combination of automated high-throughput sample handling, together with miniaturized liquid chromatography approaches and powerful mass spectrometric technologies, will greatly facilitate the identification and characterization of bioactive endogenous peptides in future studies.
Endogenous peptides in the clinic
There are several disadvantages that limit the use of peptides as therapeutic agents, including the fact they are usually not orally available and are often unstable. Nonetheless, they are already used in the treatment of cancer, diabetes and cardiovascular diseases28,29. There is a growing interest in using peptides in other therapeutic areas, particularly as antimicrobial agents, owing to the increasing number of multidrug-resistant bacteria and viruses. Many antimicrobial peptides are broadly active, as they often target conserved components of bacterial membranes15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30. Furthermore, such peptides neutralize endotoxins and it is difficult for most pathogens to develop resistance81; for example, HIV-1 variants only showed reduced sensitivity to VIRIP after more than 1 year of cell culture passage in the presence of the inhibitor82. Accumulating evidence also suggests that antimicrobial peptides have immunomodulatory and antineoplastic properties and thus offer prospects for the treatment of inflammation and cancer83,84,85. Other potential advantages of peptides are that they are usually easy to synthesize and can be readily modified to optimize their activity, specificity and stability. Notably, the costs of peptide syntheses are rapidly decreasing and novel delivery methods may increase their efficacy and eventually even enable oral application. As a consequence, the number of peptidic compounds that are entering clinical trials is steadily increasing and many candidates are in clinical and preclinical development29. Thus, although low stability under physiological conditions and lack of oral bioavailability are still important challenges26,27,28,29,30,31, the strategy to optimize bioactive endogenous peptides for clinical applications seems to be highly promising. Alternatively, it may be possible to induce or apply the proteases that generate the active peptides from their abundant precursors to treat infectious or inflammatory diseases.
Conclusions and perspectives
The human peptidome probably contains many unknown factors that may have key roles in antimicrobial immunity and inflammatory processes. These bioactive peptides cannot be identified by genomics and transcriptomics approaches, as they do not detect post-translational modifications. Peptide libraries that are derived from bodily fluids or tissues will facilitate the systematic and unbiased identification of these 'hidden treasures'. Some activities may certainly be missed because they are relatively weak or because some antimicrobial agents may be degraded or inactivated during sample processing or storage. Nonetheless, this methodology seems to be suitable to identify the most potent and stable endogenous antimicrobial peptides. In the longer term, we envision the highly reproducible generation of peptide libraries from multiple sources in combination with robust assays for the identification of factors that affect key pathogens and biological activities. We also speculate that novel powerful techniques for the purification, optimization, production and delivery of endogenous peptides will greatly facilitate their future clinical development.
References
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Jensen, O. N. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 8, 33–41 (2004).
Petricoin, E. F., Belluco, C., Araujo, R. P. & Liotta, L. A. The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nature Rev. Cancer 6, 961–967 (2006).
Zucht, H. D., Raida, M., Adermann, K., Mägert, H. J. & Forssmann, W. G. Casocidin-I: a casein-αs2 derived peptide exhibits antibacterial activity. FEBS Lett. 372, 185–188 (1995).
Mägert, H. J. et al. LEKTI, a novel 15-domain type of human serine proteinase inhibitor. J. Biol. Chem. 274, 21499–21502 (1999).
Detheux, M. et al. Natural proteolytic processing of hemofiltrate CC chemokine 1 generates a potent CC chemokine receptor (CCR)1 and CCR5 agonist with anti-HIV properties. J. Exp. Med. 192, 1501–1508 (2000).
Liepke, C. et al. Human hemoglobin-derived peptides exhibit antimicrobial activity: a class of host defense peptides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 791, 345–356 (2003).
Münch, J. et al. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell 129, 263–275 (2007).
Påhlman, L. I. et al. Antimicrobial activity of fibrinogen and fibrinogen-derived peptides — a novel link between coagulation and innate immunity. Thromb. Haemost. 109, 930–939 (2013).
Papareddy, P., Mörgelin, M., Walse, B., Schmidtchen, A. & Malmsten, M. Antimicrobial activity of peptides derived from human ß-amyloid precursor protein. J. Pept. Sci. 18, 183–191 (2012).
Papareddy, P. et al. C-terminal peptides of tissue factor pathway inhibitor are novel host defense molecules. J. Biol. Chem. 285, 28387–28398 (2010).
Kalle, M. et al. Proteolytic activation transforms heparin cofactor II into a host defense molecule. J. Immunol. 190, 6303–6310 (2013).
Wagener, J. et al. A peptide derived from the highly conserved protein GAPDH is involved in tissue protection by different antifungal strategies and epithelial immunomodulation. J. Invest. Dermatol. 133, 144–153 (2013).
Yount, N. Y. et al. Context mediates antimicrobial efficacy of kinocidin congener peptide RP-1. PLoS ONE 6, e26727 (2011).
Choi, K. Y., Chow, L. N. & Mookherjee, N. Cationic host defence peptides: multifaceted role in immune modulation and inflammation. J. Innate Immun. 4, 361–370 (2012).
Guaní-Guerra, E., Santos-Mendoza, T., Lugo-Reyes, S. O. & Terán, L. M. Antimicrobial peptides: general overview and clinical implications in human health and disease. Clin. Immunol. 135, 1–11 (2010).
Ding, J., Chou, Y. Y. & Chang, T. L. Defensins in viral infections. J. Innate Immun. 1, 413–420 (2009).
Bahar, A. A. & Ren, D. Antimicrobial peptides. Pharmaceuticals 6, 1543–1575 (2013).
Ganz, T. et al. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Invest. 76, 1427–1435 (1985).
Harder, J., Bartels, J., Christophers, E. & Schroder, J. M. Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276, 5707–5713 (2001).
Oudhoff, M. J. et al. Histatins are the major wound-closure stimulating factors in human saliva as identified in a cell culture assay. FASEB J. 22, 3805–3812 (2008).
Fleming, A. On a remarkable bacteriolytic element found in tissues and secretions. Proc. R. Soc. Lond. B. 93, 306–317 (1922).
Andersson, E. et al. Isolation of human cationic antimicrobial protein-18 from seminal plasma and its association with prostasomes. Hum. Reprod. 17, 2529–2534 (2002).
Malm, J. et al. The human cationic antimicrobial protein (hCAP-18) is expressed in the epithelium of human epididymis, is present in seminal plasma at high concentrations, and is attached to spermatozoa. Infect. Immun. 68, 4297–4302 (2000).
Schittek, B. et al. Dermicidin: a novel human antibiotic peptide secreted by sweat glands. Nature Immunol. 2, 1133–1137 (2001).
Peters, B. M., Shirtliff, M. E. & Jabra-Rizk, M. A. Antimicrobial peptides: primeval molecules or future drugs? PLoS Pathog. 6, e1001067 (2010).
Prado Montes de Oca, E. Antimicrobial peptide elicitors: new hope for the post-antibiotic era. Innate Immun. 19, 227–241 (2013).
Fox, J. L. Antimicrobial peptides stage a comeback. Nature Biotech. 31, 379–382 (2013).
Craik, D. J., Fairlie, D. P., Liras, S. & Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 81, 136–147 (2013).
Yount, N. Y. & Yeaman, M. R. Emerging themes and therapeutic prospects for anti-infective peptides. Annu. Rev. Pharmacol. Toxicol. 52, 337–360 (2012).
Sun, L. Peptide-based drug development. Mod. Chem. Appl. 1, e103 (2013).
Schulz-Knappe, P., Raida, M., Meyer, M., Quellhorst, E. A. & Forssmann, W. G. Systematic isolation of circulating human peptides: the concept of peptide trapping. Eur. J. Med. Res. 1, 223–236 (1996).
Schulz-Knappe, P. et al. Peptide bank generated by large-scale preparation of circulating human peptides. J. Chromatogr. A 776, 125–132 (1997).
Richter, R. et al. Composition of the peptide fraction in human blood plasma: database of circulating human peptides. J. Chromatogr. B Biomed. Sci. Appl. 726, 25–35 (1999).
Raida, M., Schulz-Knappe, P., Heine, G. & Forssmann, W. G. Liquid chromatography and electrospray mass spectrometric mapping of peptides from human plasma filtrate. J. Am. Soc. Mass Spectrom. 10, 45–54 (1999).
Jiang, S., Lin, K., Strick, N. & Neurath, A. R. HIV-1 inhibition by a peptide. Nature 365, 113 (1993).
He, Y. Synthesized peptide inhibitors of HIV-1 gp41-dependent membrane fusion. Curr. Pharm. Des. 19, 1800–1809 (2013).
Cocchi, F. et al. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science 270, 1811–1815 (1995).
Deng, H. et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381, 661–666 (1996).
Gaertner, H. et al. Highly potent, fully recombinant anti-HIV chemokines: reengineering a low-cost microbicide. Proc. Natl Acad. Sci. USA 105, 17706–17711 (2008).
Harris, R. S., Hultquist, J. F. & Evans, D. T. The restriction factors of human immunodeficiency virus. J. Biol. Chem. 287, 40875–40883 (2012).
Münch, J. et al. Hemofiltrate CC chemokine 1[9-74] causes effective internalization of CCR5 and is a potent inhibitor of R5-tropic human immunodeficiency virus type 1 strains in primary T cells and macrophages. Antimicrob. Agents Chemother. 46, 982–990 (2002).
Auerbach, D. J. et al. Identification of the platelet-derived chemokine CXCL4/PF-4 as a broad-spectrum HIV-1 inhibitor. Proc. Natl Acad. Sci. USA 109, 9569–9574 (2012).
McMorran, B. J. et al. Platelet factor 4 and Duffy antigen required for platelet killing of Plasmodium falciparum. Science 338, 1348–1351 (2012).
Blumenthal, R. & Dimitrov, D. S. Targeting the sticky fingers of HIV-1. Cell 129, 243–245 (2007).
Harter, C., James, P., Bächi, T., Semenza, G. & Brunner, J. Hydrophobic binding of the ectodomain of influenza hemagglutinin to membranes occurs through the 'fusion peptide'. J. Biol. Chem. 264, 6459–6464 (1989).
Drummer, H. E., Boo, I. & Poumbourios, P. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J. Gen. Virol. 88, 1144–1148 (2007).
Madu, I. G., Roth, S. L., Belouzard, S. & Whittaker, G. R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 83, 7411–7421 (2009).
Glushakova, S. E., Lukashevich, I. S. & Baratova, L. A. Prediction of arenavirus fusion peptides on the basis of computer analysis of envelope protein sequences. FEBS Lett. 269, 145–147 (1990).
Rodríguez-Crespo, I. et al. Structural properties of the putative fusion peptide of hepatitis B virus upon interaction with phospholipids. Circular dichroism and Fourier-transform infrared spectroscopy studies. Eur. J. Biochem. 242, 243–248 (1996).
Kramer, H. B. et al. Elevation of intact and proteolytic fragments of acute phase proteins constitutes the earliest systemic antiviral response in HIV-1 infection. PLoS Pathog. 6, e1000893 (2010).
Collin, M. et al. Constitutive expression of the antibacterial CXC chemokine GCP-2/CXCL6 by epithelial cells of the male reproductive tract. J. Reprod. Immunol. 79, 37–43 (2008).
Edström, A. M. et al. The major bactericidal activity of human seminal plasma is zinc-dependent and derived from fragmentation of the semenogelins. J. Immunol. 181, 3413–3421 (2008).
Linge, H. M. et al. The antibacterial chemokine MIG/CXCL9 is constitutively expressed in epithelial cells of the male urogenital tract and is present in seminal plasma. J. Interferon Cytokine Res. 28, 191–196 (2008).
Yenugu, S. et al. Antibacterial properties of the sperm-binding proteins and peptides of human epididymis 2 (HE2) family; salt sensitivity, structural dependence and their interaction with outer and cytoplasmic membranes of Escherichia coli. Biochem. J. 372, 473–483 (2003).
Martellini, J. A. et al. Cationic polypeptides contribute to the anti-HIV-1 activity of human seminal plasma. FASEB J. 23, 3609–3618 (2009).
Münch, J. et al. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell 131, 1059–1071 (2007).
Münch, J. et al. Effect of semen and seminal amyloid on vaginal transmission of simian immunodeficiency virus. Retrovirology 10, 148 (2013).
Roan, N. R. et al. The cationic properties of SEVI underlie its ability to enhance human immunodeficiency virus infection. J. Virol. 83, 73–80 (2009).
Arnold, F. et al. Naturally occurring fragments from two distinct regions of the prostatic acid phosphatase form amyloidogenic enhancers of HIV infection. J. Virol. 86, 1244–1249 (2012).
Roan, N. R. et al. Peptides released by physiological cleavage of semen coagulum proteins form amyloids that enhance HIV infection. Cell Host Microbe 10, 541–550 (2011).
Usmani, S. M. et al. Direct visualization of HIV-enhancing endogenous amyloid fibrils in human semen. Nature Commun. 5, 3508 (2014).
Kim, K. A. et al. Semen-mediated enhancement of HIV infection is donor-dependent and correlates with the levels of SEVI. Retrovirology 7, 55 (2010).
Capule, C. C., Brown, C., Olsen, J. S., Dewhurst, S. & Yang, J. Oligovalent amyloid-binding agents reduce SEVI-mediated enhancement of HIV-1 infection. J. Am. Chem. Soc. 134, 905–908 (2012).
Hauber, I., Hohenberg, H., Holstermann, B., Hunstein, W. & Hauber, J. The main green tea polyphenol epigallocatechin-3-gallate counteracts semen-mediated enhancement of HIV infection. Proc. Natl Acad. Sci. USA 106, 9033–9038 (2009).
Olsen, J. S. et al. Amyloid-binding small molecules efficiently block SEVI (semen-derived enhancer of virus infection) and semen-mediated enhancement of HIV-1 infection. J. Biol. Chem. 285, 35488–35496 (2010).
Roan, N. R., Sowinski, S., Münch, J., Kirchhoff, F. & Greene, W. C. Aminoquinoline surfen inhibits the action of SEVI (semen-derived enhancer of viral infection). J. Biol. Chem. 285, 1861–1869 (2010).
Sievers, S. A. et al. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 475, 96–100 (2011).
Tang, Q., Roan, N. R. & Yamamura, Y. Seminal plasma and semen amyloids enhance cytomegalovirus infection in cell culture. J. Virol. 87, 12583–12591 (2013).
Easterhoff, D. et al. Semen-derived enhancer of viral infection (SEVI) binds bacteria, enhances bacterial phagocytosis by macrophages, and can protect against vaginal infection by a sexually transmitted bacterial pathogen. Antimicrob. Agents Chemother. 57, 2443–2450 (2013).
Wong, C. C., Cheng, K. W. & Rigas, B. Preclinical predictors of anticancer drug efficacy: critical assessment with emphasis on whether nanomolar potency should be required of candidate agents. J. Pharmacol. Exp. Ther. 341, 572–578 (2012).
Forssmann, U. et al. n-Nonanoyl-CC chemokine ligand 14, a potent CC chemokine ligand 14 analogue that prevents the recruitment of eosinophils in allergic airway inflammation. J. Immunol. 173, 3456–3466 (2004).
Forssmann, W. G. et al. Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide. Sci. Transl. Med. 2, 63re3 (2010).
Wurm, M. et al. The influence of semen-derived enhancer of virus infection on the efficiency of retroviral gene transfer. J. Gene Med. 12, 137–146 (2010).
Yolamanova, M. et al. Peptide nanofibrils boost retroviral gene transfer and provide a rapid means for concentrating viruses. Nature Nanotechnol. 8, 130–136 (2013).
Rajamäki, K. et al. Extracellular acidosis is a novel danger signal alerting innate immunity via the NLRP3 inflammasome. J. Biol. Chem. 288, 13410–13419 (2013).
Okajima, F. Regulation of inflammation by extracellular acidification and proton-sensing GPCRs. Cell. Signal. 25, 2263–2271 (2013).
Bensch, K. W., Raida, M., Mägert, H. J., Schulz-Knappe, P. & Forssmann, W. G. hBD-1: a novel β-defensin from human plasma. FEBS Lett. 368, 331–335 (1995).
Borst, E. M. et al. A peptide inhibitor of cytomegalovirus infection from human hemofiltrate. Antimicrob. Agents Chemother. 57, 4751–4560 (2013).
Angel, T. E. et al. Mass spectrometry-based proteomics: existing capabilities and future directions. Chem. Soc. Rev. 41, 3912–3928 (2012).
Marr, A. K., Gooderham, W. J. & Hancock, R. E. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr. Opin. Pharmacol. 6, 468–472 (2006).
Gonzalez, E., Ballana, E., Clotet, B. & Esté, J. A. Development of resistance to VIR-353 with cross-resistance to the natural HIV-1 entry virus inhibitory peptide (VIRIP). AIDS 25, 1557–1583 (2011).
Gaspar, D., Veiga, A. S. & Castanho, M. A. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 4, 294 (2013).
Chow, J. Y., Li, Z. J., Wu, W. K. & Cho, C. H. Cathelicidin a potential therapeutic peptide for gastrointestinal inflammation and cancer. World J. Gastroenterol. 19, 2731–2635 (2013).
Hilchie, A. L., Wuerth, K. & Hancock, R. E. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nature Chem. Biol. 9, 761–768 (2013).
Krause, A. et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 480, 147–150 (2000).
Ganz, T. & Lehrer, R. I. Defensins. Pharmacol. Ther. 66, 191–205 (1995).
Krause, A. et al. Human natriuretic peptides exhibit antimicrobial activity. Eur. J. Med. Res. 6, 215–218 (2001).
Liepke, C., Zucht, H. D., Forssmann, W. G. & Ständker, L. Purification of novel peptide antibiotics from human milk. J. Chromatogr. B Biomed. Sci. Appl. 752, 369–377 (2001).
Ständker, L. et al. Quantitative enzyme-linked immunosorbent assay determination of an abundant haemoglobin derived anti-infective peptide in human placenta. Anal. Biochem. 401, 53–60 (2010).
Albericio, F. & Kruger, H. G. Therapeutic peptides. Future Med. Chem. 4, 1527–1531 (2012).
Acknowledgements
The authors thank current and past members of their laboratories as well as all of their collaborators for their contributions to the work that is discussed in this article. They apologize to the authors of the many interesting studies that could not be cited owing to space limitations. The authors are supported by grants from the Deutsche Forschungsgemeinschaft, the Volkswagenstiftung, the German Federal Ministry of Education and Research (BMBF), the European Community and the European Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Münch, J., Ständker, L., Forssmann, WG. et al. Discovery of modulators of HIV-1 infection from the human peptidome. Nat Rev Microbiol 12, 715–722 (2014). https://doi.org/10.1038/nrmicro3312
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3312
This article is cited by
-
Some novel peptides containing a modified pyrazolopyrimidine moiety: design, synthesis, and in vitro antibacterial screening
Applied Biological Chemistry (2023)
-
CXCR4 in Waldenström’s Macroglobulinema: chances and challenges
Leukemia (2021)
-
Alpha-1 antitrypsin inhibits TMPRSS2 protease activity and SARS-CoV-2 infection
Nature Communications (2021)
-
Digging into the low molecular weight peptidome with the OligoNet web server
Scientific Reports (2017)
-
The KT Jeang Retrovirology prize 2016: Frank Kirchhoff
Retrovirology (2016)