Key Points
-
Hepatitis C virus (HCV) is an enveloped, positive-strand RNA virus. Unlike other enveloped RNA viruses, HCV particles interact with serum lipoproteins, which are important for the infectivity of HCV particles.
-
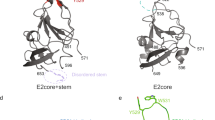
New structural information is available for E2 glycoprotein of the related pestiviruses. Common features of HCV and pestiviruses suggest that these viruses use an uncharacterized mechanism of viral fusion.
-
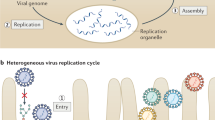
HCV particles enter cells via a multistep process involving numerous cell surface proteins, cellular processing of virus-associated lipoproteins, signal transduction events, clathrin-mediated internalization of the virus, and low-pH-induced membrane fusion.
-
HCV particle assembly occurs via budding into the ER. This process requires recently defined interactions between the viral structural and non-structural proteins.
-
Nascent HCV particles exit the cell via transit through the secretory pathway. During this process, HCV undergoes maturational events similar to those of serum lipoproteins.
Abstract
Hepatitis C virus, a major human pathogen, produces infectious virus particles with several unique features, such as an ability to interact with serum lipoproteins, a dizzyingly complicated process of virus entry, and a pathway of virus assembly and release that is closely linked to lipoprotein secretion. Here, we review these unique features, with an emphasis on recent discoveries concerning virus particle structure, virus entry and virus particle assembly and release.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lavanchy, D. The global burden of hepatitis C. Liver Int. 29 (Suppl. 1), 74–81 (2009).
Pawlotsky, J. M. Treatment of chronic hepatitis C: current and future. Curr. Top. Microbiol. Immunol. 369, 321–342 (2013).
Lindenbach, B. D., Murray, C. L., Thiel, H. J. & Rice, C. M. in Fields Virology (eds Knipe, D. M. & Howley, P. M.) 712–746 (Lippincott Williams & Wilkins, 2013).
King, A. M. Q., Lefkowitz, E., Adams, M. J. & Carstens, E. B. (eds) Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses (Elsevier, 2011).
Morikawa, K. et al. Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus. J. Viral Hepat 18, 305–315 (2011).
Lohmann, V. Hepatitis C virus RNA replication. Curr. Top. Microbiol. Immunol. 369, 167–198 (2013).
Niepmann, M. Hepatitis C virus RNA translation. Curr. Top. Microbiol. Immunol. 369, 143–166 (2013).
Schoggins, J. W. & Rice, C. M. Innate immune responses to hepatitis C virus. Curr. Top. Microbiol. Immunol. 369, 219–242 (2013).
Feinstone, S. M., Kapikian, A. Z., Purcell, R. H., Alter, H. J. & Holland, P. V. Transfusion-associated hepatitis not due to viral hepatitis type A or B. N. Engl. J. Med. 292, 767–770 (1975).
Choo, Q. L. et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244, 359–362 (1989). This is a classic paper that identifies HCV as the aetiological agent of non-A, non-B hepatitis.
Kolykhalov, A. A. et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277, 570–574 (1997).
Yanagi, M., Purcell, R. H., Emerson, S. U. & Bukh, J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl Acad. Sci. USA 94, 8738–8743 (1997).
Lohmann, V. HCV replicons: overview and basic protocols. Methods Mol. Biol. 510, 145–163 (2009).
Baumert, T. F., Ito, S., Wong, D. T. & Liang, T. J. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72, 3827–3836 (1998).
Bartosch, B., Dubuisson, J. & Cosset, F. L. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J. Exp. Med. 197, 633–642 (2003).
Drummer, H. E., Maerz, A. & Poumbourios, P. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546, 385–390 (2003).
Hsu, M. et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl Acad. Sci. USA 100, 7271–7276 (2003).
Lindenbach, B. D. et al. Complete replication of hepatitis C virus in cell culture. Science 309, 623–626 (2005).
Wakita, T. et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature Med. 11, 791–796 (2005).
Zhong, J. et al. Robust hepatitis C virus infection in vitro. Proc. Natl Acad. Sci. USA 102, 9294–9299 (2005). References 18–20 describe the first HCVcc systems.
Steinmann, E. & Pietschmann, T. Cell culture systems for hepatitis C virus. Curr. Top. Microbiol. Immunol. 369, 17–48 (2013).
Yamamoto, T., Takahashi, S., Moriwaki, Y., Hada, T. & Higashino, K. A newly discovered apolipoprotein B- containing high-density lipoprotein produced by human hepatoma cells. Biochim. Biophys. Acta 922, 177–183 (1987).
Lindenbach, B. D. et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl Acad. Sci. USA 103, 3805–3809 (2006).
Podevin, P. et al. Production of infectious hepatitis C virus in primary cultures of human adult hepatocytes. Gastroenterology 139, 1355–1364 (2010).
Ploss, A. et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc. Natl Acad. Sci. USA 107, 3141–3145 (2010).
Billerbeck, E., de Jong, Y., Dorner, M., de la Fuente, C. & Ploss, A. Animal models for hepatitis C. Curr. Top. Microbiol. Immunol. 369, 49–86 (2013).
Tscherne, D. M. et al. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80, 1734–1741 (2006).
Bradley, D. W. et al. Posttransfusion non-A, non-B hepatitis in chimpanzees: physicochemical evidence that the tubule-forming agent is a small, enveloped virus. Gastroenterology 88, 773–779 (1985).
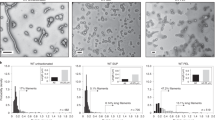
Gastaminza, P. et al. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 84, 10999–11009 (2010). This study provides high-resolution images of highly enriched HCVcc particles.
He, L. F. et al. Determining the size of non-A, non-B hepatitis virus by filtration. J. Infect. Dis. 156, 636–640 (1987).
Merz, A. et al. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 286, 3018–3032 (2011). This report describes proteomic, lipidomic and structural analyses of highly purified HCVcc particles.
Catanese, M. T. et al. Ultrastructural analysis of hepatitis C virus particles. Proc. Natl Acad. Sci. USA 110, 9505–9510 (2013). This work obtains high-resolution images of affinity-captured HCVcc particles.
Mukhopadhyay, S., Kuhn, R. J. & Rossmann, M. G. A structural perspective of the flavivirus life cycle. Nature Rev. Microbiol. 3, 13–22 (2005).
Modis, Y., Ogata, S., Clements, D. & Harrison, S. C. Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 (2004).
DuBois, R. M. et al. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature 493, 552–556 (2013).
Lescar, J. et al. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105, 137–148 (2001).
Dessau, M. & Modis, Y. Crystal structure of glycoprotein C from Rift Valley fever virus. Proc. Natl Acad. Sci. USA 110, 1696–1701 (2013).
Kielian, M. & Rey, F. A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nature Rev. Microbiol. 4, 67–76 (2006).
Kadlec, J., Loureiro, S., Abrescia, N. G., Stuart, D. I. & Jones, I. M. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nature Struct. Mol. Biol. 15, 1024–1030 (2008).
Garry, R. F. & Dash, S. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology 307, 255–265 (2003).
Krey, T. et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 6, e1000762 (2010).
Yagnik, A. T. et al. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40, 355–366 (2000).
El Omari, K., Iourin, O., Harlos, K., Grimes, J. M. & Stuart, D. I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 3, 30–35 (2013).
Li, Y., Wang, J., Kanai, R. & Modis, Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl Acad. Sci. USA 110, 6805–6810 (2013). This article reports the crystal structure of a pestivirus E2 glycoprotein, revealing a new viral glycoprotein architecture that might serve as a model for HCV E2.
Yu, I. M. et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 (2008).
Pileri, P. et al. Binding of hepatitis C virus to CD81. Science 282, 938–941 (1998).
Hulst, M. M. & Moormann, R. J. Inhibition of pestivirus infection in cell culture by envelope proteins Erns and E2 of classical swine fever virus: Erns and E2 interact with different receptors. J. Gen. Virol. 78, 2779–2787 (1997).
Scarselli, E. et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21, 5017–5025 (2002).
Krey, T., Thiel, H. J. & Rümenapf, T. Acid-resistant bovine pestivirus requires activation for pH-triggered fusion during entry. J. Virol. 79, 4191–4200 (2005).
Vieyres, G. et al. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J. Virol. 84, 10159–10168 (2010).
Drummer, H. E., Boo, I. & Poumbourios, P. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J. Gen. Virol. 88, 1144–1148 (2007).
Flint, M. et al. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J. Virol. 73, 6782–6790 (1999).
Hijikata, M. et al. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J. Virol. 67, 1953–1958 (1993).
Thomssen, R. et al. Association of hepatitis C virus in human sera with β-lipoprotein. Med. Microbiol. Immunol. 181, 293–300 (1992).
Lindenbach, B. D. Virion assembly and release. Curr. Top. Microbiol. Immunol. 369, 199–218 (2013).
Kono, Y., Hayashida, K., Tanaka, H., Ishibashi, H. & Harada, M. High-density lipoprotein binding rate differs greatly between genotypes 1b and 2a/2b of hepatitis C virus. J. Med. Virol. 70, 42–48 (2003).
Felmlee, D. J. et al. Intravascular transfer contributes to postprandial increase in numbers of very-low-density hepatitis C virus particles. Gastroenterology 139, 1774–1783 (2010). This study shows that the interaction between HCV and serum lipoproteins is transient and exchangeable in vivo.
Diaz, O. et al. Preferential association of hepatitis C virus with apolipoprotein B48-containing lipoproteins. J. Gen. Virol. 87, 2983–2991 (2006).
Chang, K. S., Jiang, J., Cai, Z. & Luo, G. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81, 13783–13793 (2007).
Meunier, J. C. et al. Apolipoprotein C1 association with hepatitis C virus. J. Virol. 82, 9647–9656 (2008).
Hsu, V. W., Bai, M. & Li, J. Getting active: protein sorting in endocytic recycling. Nature Rev. Mol. Cell Biol. 13, 323–328 (2012).
Helenius, A. in Fields Virology (eds Knipe, D. M. & Howley, P. M.) 87–104 (Lippincott Williams & Wilkins, 2013).
Agnello, V., Abel, G., Elfahal, M., Knight, G. B. & Zhang, Q. X. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl Acad. Sci. USA 96, 12766–12771 (1999).
Germi, R. et al. Cellular glycosaminoglycans and low density lipoprotein receptor are involved in hepatitis C virus adsorption. J. Med. Virol. 68, 206–215 (2002).
Monazahian, M. et al. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 57, 223–229 (1999).
Albecka, A. et al. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology 55, 998–1007 (2012).
Petracca, R. et al. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74, 4824–4830 (2000).
Bertaux, C. & Dragic, T. Different domains of CD81 mediate distinct stages of hepatitis C virus pseudoparticle entry. J. Virol. 80, 4940–4948 (2006).
Bankwitz, D. et al. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J. Virol. 84, 5751–5763 (2010).
Sharma, N. R. et al. Hepatitis C virus is primed by CD81 protein for low pH-dependent fusion. J. Biol. Chem. 286, 30361–30376 (2011).
Harris, H. J. et al. Claudin association with CD81 defines hepatitis C virus entry. J. Biol. Chem. 285, 21092–21102 (2010).
Dorner, M. et al. A genetically humanized mouse model for hepatitis C virus infection. Nature 474, 208–211 (2011).
Dorner, M. et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature http://dx.doi.org/10.1038/nature12427 (2013). References 72 and 73 demonstrate that mice expressing human CD81 and OCLN are permissive to HCV infection.
Acton, S. et al. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271, 518–520 (1996).
Dao Thi, V. L. et al. Characterization of hepatitis C virus particle subpopulations reveals multiple usage of the scavenger receptor BI for entry steps. J. Biol. Chem. 287, 31242–31257 (2012).
Zahid, M. N. et al. The postbinding activity of scavenger receptor class B type I mediates initiation of hepatitis C virus infection and viral dissemination. Hepatology 57, 492–504 (2013).
Evans, M. J. et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446, 801–805 (2007).
Reynolds, G. M. et al. Hepatitis C virus receptor expression in normal and diseased liver tissue. Hepatology 47, 418–427 (2008).
Ploss, A. et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457, 882–886 (2009).
Sourisseau, M. et al. Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies. PLoS Pathog. 9, e1003244 (2013).
Martin, D. N. & Uprichard, S. L. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc. Natl Acad. Sci. USA (2013).
Sainz, B. Jr et al. Identification of the Niemann-Pick C1- like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nature Med. 18, 281–285 (2012).
Jia, L., Betters, J. L. & Yu, L. Niemann-pick C1-like 1 (NPC1L1) protein in intestinal and hepatic cholesterol transport. Annu. Rev. Physiol. 73, 239–259 (2011).
Farquhar, M. J. et al. Protein kinase A-dependent step(s) in hepatitis C virus entry and infectivity. J. Virol. 82, 8797–8811 (2008).
Brazzoli, M. et al. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 82, 8316–8329 (2008).
Farquhar, M. J. et al. Hepatitis C virus induces CD81 and claudin-1 endocytosis. J. Virol. 86, 4305–4316 (2012).
Lupberger, J. et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nature Med. 17, 589–595 (2011). This investigation finds that EGFR and EPHA2 are essential for HCV entry because of their ability to promote the CD81–CLDN1 interaction.
Diao, J. et al. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J. Virol. 86, 10935–10949 (2012).
Zona, L. et al. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe 13, 302–313 (2013).
Lupberger, J. et al. EGFR signaling impairs the antiviral activity of interferon-alpha. Hepatology http://dx.doi.org/10.1002/hep.26404 (2013).
Coller, K. E. et al. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 5, e1000702 (2009).
Sattentau, Q. Avoiding the void: cell-to-cell spread of human viruses. Nature Rev. Microbiol. 6, 815–826 (2008).
Wahid, A. & Dubuisson, J. Virus-neutralizing antibodies to hepatitis C virus. J. Viral Hepat. 20, 369–376 (2013).
Brimacombe, C. L. et al. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J. Virol. 85, 596–605 (2011).
Catanese, M. T. et al. Different requirements for SR-BI in hepatitis C virus cell-free versus cell-to-cell transmission. J. Virol. 87, 8282–8293 (2013).
Timpe, J. M. et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47, 17–24 (2008).
Witteveldt, J. et al. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 90, 48–58 (2009).
Gastaminza, P. et al. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82, 2120–2129 (2008).
Gastaminza, P., Kapadia, S. B. & Chisari, F. V. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80, 11074–11081 (2006).
Boulant, S. et al. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281, 22236–22247 (2006).
Majeau, N. et al. Palmitoylation of hepatitis C virus core protein is important for virion production. J. Biol. Chem. 284, 33915–33925 (2009).
Barba, G. et al. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl Acad. Sci. USA 94, 1200–1205 (1997).
Boulant, S., Vanbelle, C., Ebel, C., Penin, F. & Lavergne, J. P. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 79, 11353–11365 (2005).
Moradpour, D., Englert, C., Wakita, T. & Wands, J. R. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology 222, 51–63 (1996).
Boulant, S., Targett-Adams, P. & McLauchlan, J. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88, 2204–2213 (2007).
Miyanari, Y. et al. The lipid droplet is an important organelle for hepatitis C virus production. Nature Cell Biol. 9, 1089–1097 (2007). This study shows that trafficking of HCV core protein to LDs is important for virus particle assembly.
Shavinskaya, A., Boulant, S., Penin, F., McLauchlan, J. & Bartenschlager, R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282, 37158–37169 (2007).
Menzel, N. et al. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 8, e1002829 (2012).
Herker, E. et al. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nature Med. 16, 1295–1298 (2010).
Neveu, G. et al. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 8, e1002845 (2012).
Counihan, N. A., Rawlinson, S. M. & Lindenbach, B. D. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog. 7, e1002302 (2011). This work involves live cell imaging of functional core protein in virus-producing cells and shows that the interaction between NS2 and NS3-4A is important for recruiting the core protein from LDs into virus assembly.
Coller, K. E. et al. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 8, e1002466 (2012).
Dubuisson, J. et al. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68, 6147–6160 (1994).
Michalak, J. P. et al. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 78, 2299–2306 (1997).
Brazzoli, M. et al. Folding and dimerization of hepatitis C virus E1 and E2 glycoproteins in stably transfected CHO cells. Virology 332, 438–453 (2005).
Flint, M. et al. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74, 702–709 (2000).
Flint, M. et al. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73, 6235–6244 (1999).
Heile, J. M. et al. Evaluation of hepatitis C virus glycoprotein E2 for vaccine design: an endoplasmic reticulum-retained recombinant protein is superior to secreted recombinant protein and DNA-based vaccine candidates. J. Virol. 74, 6885–6892 (2000).
Whidby, J. et al. Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J. Virol. 83, 11078–11089 (2009).
Appel, N. et al. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4, e1000035 (2008).
Masaki, T. et al. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82, 7964–7976 (2008).
Camus, G. et al. Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J. Biol. Chem. 288, 9915–9923 (2013).
Kim, S., Welsch, C., Yi, M. & Lemon, S. M. Regulation of the production of infectious genotype 1a hepatitis C virus by NS5A domain III. J. Virol. 85, 6645–6656 (2011).
Tellinghuisen, T. L., Foss, K. L. & Treadaway, J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4, e1000032 (2008).
Benga, W. J. et al. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology 51, 43–53 (2010).
Evans, M. J., Rice, C. M. & Goff, S. P. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl Acad. Sci. USA 101, 13038–13043 (2004).
Backes, P. et al. Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 84, 5775–5789 (2010).
Jirasko, V. et al. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 6, e1001233 (2010).
Popescu, C. I. et al. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 7, e1001278 (2011).
Gentzsch, J. et al. Hepatitis C virus p7 is critical for capsid assembly and envelopment. PLoS Pathog. 9, e1003355 (2013).
Moradpour, D. & Penin, F. Hepatitis C virus proteins: from structure to function. Curr. Top. Microbiol. Immunol. 369, 113–142 (2013).
Jirasko, V. et al. Structural and functional characterization of non-structural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283, 28546–28562 (2008).
Jones, C. T., Murray, C. L., Eastman, D. K., Tassello, J. & Rice, C. M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81, 8374–8383 (2007).
Beran, R. K., Lindenbach, B. D. & Pyle, A. M. The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase. J. Virol. 83, 3268–3275 (2009).
Kuang, W. F. et al. Hepatitis C virus NS3 RNA helicase activity is modulated by the two domains of NS3 and NS4A. Biochem. Biophys. Res. Commun. 317, 211–217 (2004).
Phan, T., Kohlway, A., Dimberu, P., Pyle, A. M. & Lindenbach, B. D. The acidic domain of hepatitis C virus NS4A contributes to RNA replication and virus particle assembly. J. Virol. 85, 1193–1204 (2011).
Pietschmann, T. et al. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 5, e1000475 (2009).
Ma, Y., Yates, J., Liang, Y., Lemon, S. M. & Yi, M. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82, 7624–7639 (2008).
Phan, T., Beran, R. K., Peters, C., Lorenz, I. C. & Lindenbach, B. D. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 83, 8379–8395 (2009).
Jones, D. M., Atoom, A. M., Zhang, X., Kottilil, S. & Russell, R. S. A genetic interaction between the core and NS3 proteins of hepatitis C virus is essential for production of infectious virus. J. Virol. 85, 12351–12361 (2011).
Welsch, S., Muller, B. & Krausslich, H. G. More than one door — budding of enveloped viruses through cellular membranes. FEBS Lett. 581, 2089–2097 (2007).
Ariumi, Y. et al. The ESCRT system is required for hepatitis C virus production. PLoS ONE 6, e14517 (2011).
Corless, L., Crump, C. M., Griffin, S. D. & Harris, M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J. Gen. Virol. 91, 362–372 (2010).
Tamai, K. et al. Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology 422, 377–385 (2012).
Lai, C. K., Jeng, K. S., Machida, K. & Lai, M. M. Hepatitis C virus egress and release depend on endosomal trafficking of core protein. J. Virol. 84, 11590–11598 (2010).
Huang, H. et al. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl Acad. Sci. USA 104, 5848–5853 (2007).
Jiang, J. & Luo, G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 83, 12680–12691 (2009). This study demonstrates that apoE, but not apoB, is essential for HCV particle assembly. The association of NS5A with HCVcc particles was later called into question (see reference 31).
Nahmias, Y. et al. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology 47, 1437–1445 (2008).
Da Costa, D. et al. Reconstitution of the entire hepatitis C virus life cycle in nonhepatic cells. J. Virol. 86, 11919–11925 (2012).
Long, G. et al. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology 141, 1057–1066 (2011).
Clarke, D. et al. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 281, 37057–37068 (2006).
Griffin, S. D. et al. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 535, 34–38 (2003).
Luik, P. et al. The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc. Natl Acad. Sci. USA 106, 12712–12716 (2009).
Wozniak, A. L. et al. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 6, e1001087 (2010). This research defines two essential roles for p7 and shows that the ion channel activity of this protein is required during virus egress.
Ouyang, B. et al. Unusual architecture of the p7 channel from hepatitis C virus. Nature 498, 521–525 (2013).
de la Fuente, C., Goodman, Z. & Rice, C. M. Genetic and functional characterization of the N-terminal region of the hepatitis C virus NS2 protein. J. Virol. 87, 4130–4145 (2013).
Parhofer, K. G., Hugh, P., Barrett, R., Bier, D. M. & Schonfeld, G. Determination of kinetic parameters of apolipoprotein B metabolism using amino acids labeled with stable isotopes. J. Lipid Res. 32, 1311–1323 (1991).
Lund-Katz, S. & Phillips, M. C. High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell. Biochem. 51, 183–227 (2010).
Olofsson, S. O., Stillemark-Billton, P. & Asp, L. Intracellular assembly of VLDL: two major steps in separate cell compartments. Trends Cardiovasc. Med. 10, 338–345 (2000).
Lehner, R., Lian, J. & Quiroga, A. D. Lumenal lipid metabolism: implications for lipoprotein assembly. Arterioscler Thromb. Vasc. Biol. 32, 1087–1093 (2012).
Cunliffe, H. R. & Rebers, P. A. The purification and concentration of hog cholera virus. Can. J. Comp. Med. 32, 486–492 (1968).
Laude, H. Nonarbo-Togaviridae: comparative hydrodynamic properties of the Pestivirus genus. Arch. Virol. 62, 347–352 (1979).
Kuhn, R. J. in Fields Virology (eds Knipe, D. M. & Howley, P. M.) 629–650 (Lippincott Williams & Wilkins, 2013).
Hobman, T. C. in Fields Virology (eds Knipe, D. M. & Howley, P. M.) 687–711 (Lippincott Williams & Wilkins, 2013).
Snijder, E. J. & Kikkert, M. in Fields Virology (eds Knipe, D. M. & Howley, P. M.) 859–879 (Lippincott Williams & Wilkins, 2013).
Goff, S. P. in Fields Virology (eds Knipe, D. M. & Howley, P. M.) 1424–1473 (Lippincott Williams & Wilkins, 2013).
Herden, C., Briese, T., Lipkin, W. I. & Richt, J. A. in Fields Virology (eds Knipe, D. M. & Howley, P. M.) 1124–1150 (Lippincott Williams & Wilkins, 2013).
Lastra, R. & Acosta, J. M. Purification and partial characterization of maize mosaic virus. Intervirology 11, 215–220 (1979).
Neurath, A. R., Wiktor, T. J. & Koprowski, H. Density gradient centrifugation studies on rabies virus. J. Bacteriol. 92, 102–106 (1966).
Elliott, R. M. & Schmaljohn, C. S. in Fields Virology (eds Knipe, D. M. & Howley, P. M.) 1244–1282 (Lippincott Williams & Wilkins, 2013).
Stryer, L. Biochemistry (W. H. Freeman and Company, 1995).
Acknowledgements
The authors thank M. T. Catanese and Y. Modis for sharing data prior to publication and for helpful comments on the manuscript. B.D.L. is supported by grants from the US Public Health Service, National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID; grants AI076259, AI087925 and AI089826). C.M.R. is supported by NIAID (grants AI099284, AI072613, AI075099 and AI090055), the Office of the Director through the NIH Roadmap for Medical Research (grant DK085713), the National Cancer Institute (grant CA057973), The Rockefeller University Center for Clinical and Translational Science (grant UL1RR024143), the Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust, the Greenberg Medical Research Institute and the Starr Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Type I interferon
-
A class of cytokines that is induced by viral infection and interferes with viral replication. These cytokines include multiple interferon-α proteins, encoded by separate genes, as well as a single interferon-β.
- Subgenomic replicons
-
Genetic units that are capable of autonomous replication in a suitable host cell. A hepatitis C virus subgenomic replicon is a viral RNA that is less than genome length but still capable of autonomous replication.
- Pleiomorphic
-
Heterogeneous in morphology, form or shape.
- Buoyant density
-
A physical characteristic of an object, describing its tendency to float above liquids that have greater densities than the object. Thus, at equilibrium, the buoyant density of an object is equal to the density of the surrounding liquid. Buoyant densities are determined by using isopycnic ultracentrifugation.
- Tetraspanin
-
A member of a family of related cell surface proteins that have four membrane- spanning domains. Tetraspanins contain a small amino-terminal extracellular domain and a large carboxy-terminal extracellular domain that contains a conserved motif of disulphide-bonded cysteine residues.
- Tight junctions
-
Specialized plasma membrane compartments between two adjacent cells. These junctions are impermeable to small molecules and charged ions, thereby physically separating the apical and basoateral surfaces of cells. Also known as the zonula occludens.
- Secretory pathway
-
A series of subcellular membrane-bound compartments that traffic proteins and small molecules from the inside to the outside of cells.
Rights and permissions
About this article
Cite this article
Lindenbach, B., Rice, C. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol 11, 688–700 (2013). https://doi.org/10.1038/nrmicro3098
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3098
This article is cited by
-
Paired immunoglobulin-like receptor B is an entry receptor for mammalian orthoreovirus
Nature Communications (2023)
-
High dose of bile acid enables the cellular entry and replication of hepatitis C virus in vitro
Molecular & Cellular Toxicology (2022)
-
Die Virushepatitiden A bis E: Prävalenz, Erregermerkmale und Pathogenese
Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz (2022)
-
Hepatitis C virus vaccine design: focus on the humoral immune response
Journal of Biomedical Science (2020)
-
Decremental effect of non-host humoral milieu interfacing viral envelope on the RNA level and surface antigenicity of hepatitis C virus (HCV) in vitro
Molecular & Cellular Toxicology (2019)