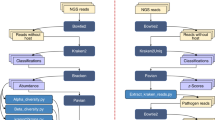
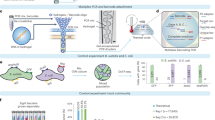
Abstract
Here, we take a snapshot of the high-throughput sequencing platforms, together with the relevant analytical tools, that are available to microbiologists in 2012, and evaluate the strengths and weaknesses of these platforms in obtaining bacterial genome sequences. We also scan the horizon of future possibilities, speculating on how the availability of sequencing that is 'too cheap to metre' might change the face of microbiology forever.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Fleischmann, R. D. et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512 (1995).
Metzker, M. L. Emerging technologies in DNA sequencing. Genome Res. 15, 1767–1776 (2005).
Venter, J. C. Multiple personal genomes await. Nature 464, 676–677 (2010).
Caruccio, N. in High-Throughput Next Generation Sequencing: Methods and Applications. Methods in Molecular Biology Vol. 733 (eds Kwon, Y. M. & Ricke, S. C.) 241–255 (Humana Press, 2011).
Bentley, D. R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Ronaghi, M., Uhlen, M. & Nyren, P. A sequencing method based on real-time pyrophosphate. Science 281 363–365 (1998).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Rothberg, J. M. et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352 (2011).
Valouev, A. et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 18, 1051–1063 (2008).
Drmanac, R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
Bowers, J. et al. Virtual terminator nucleotides for next-generation DNA sequencing. Nature Methods 6, 593–595 (2009).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Levene, M. J. et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299, 682–686 (2003).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Hess, M. et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331, 463–467 (2011).
Mutreja, A. et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477, 462–465 (2011).
Harris, S. R. et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nature Genet. 44, 413–419 (2012).
Loman, N. J. et al. Performance comparison of bench-top high-throughput sequencing platforms. Nature Biotech. 30, 434–439 (2012).
Rohde, H. et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 365, 718–724 (2011).
Mellmann, A. et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE 6, e22751 (2011).
Bashir, A. et al. A hybrid approach for the automated finishing of bacterial genomes. Nature Biotech. 1 Jul 2012 (doi:10.1038/nbt.2288).
Koren, S. et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nature Biotech. 1 Jul 2012 (doi:10.1038/nbt.2280).
Chevreux, B. et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14, 1147–1159 (2004).
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Darling, A. C., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004).
Angiuoli, S. V. & Salzberg, S. L. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27, 334–342 (2011).
Milne, I. et al. Tablet—next generation sequence assembly visualization. Bioinformatics 26, 401–402 (2010).
Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641 (1999).
Schattner, P., Brooks, A. N. & Lowe, T. M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689 (2005).
Lagesen, K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 (2007).
Aziz, R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008).
Markowitz, V. M. et al. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25, 2271–2278 (2009).
Richardson, E. J. & Watson, M. The automatic annotation of bacterial genomes. Brief. Bioinform. 9 Mar 2012 (doi:10.1093/bib/bbs007).
Chaudhuri, R. R. et al. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 36, D543–D546 (2008).
Marttinen, P. et al. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 40, e6 (2012).
Didelot, X. & Falush, D. Inference of bacterial microevolution using multilocus sequence data. Genetics 175, 1251–1266 (2007).
Didelot, X., Lawson, D., Darling, A. & Falush, D. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics 186, 1435–1449 (2010).
Domazet-Lošo, M. & Haubold, B. Alignment-free detection of local similarity among viral and bacterial genomes. Bioinformatics 27, 1466–1472 (2011).
Köser, C. U. et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366, 2267–2275 (2012).
Lewis, T. et al. High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak. J. Hosp. Infect. 75, 37–41 (2010).
Gardy, J. L. et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 364, 730–739 (2011).
Harris, S. R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474 (2011).
Beres, S. B. et al. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc. Natl Acad. Sci. USA 107, 4371–4376 (2010).
Cramer, N. et al. Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ. Microbiol. 13, 1690–1704 (2011).
Dunham, E. J. et al. Different evolutionary trajectories of European avian-like and classical swine H1N1 influenza A viruses. J. Virol. 83, 5485–5494 (2009).
Bos, K. I. et al. A draft genome of Yersinia pestis from victims of the Black Death. Nature 478, 506–510 (2011).
Woyke, T. et al. Assembling the marine metagenome, one cell at a time. PLoS ONE 4, e5299 (2009).
Lipkin, W. I. Microbe hunting. Microbiol. Mol. Biol. Rev. 74, 363–377 (2010).
Sorek, R. & Cossart, P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nature Rev. Genet. 11, 9–16 (2010).
Passalacqua, K. D. et al. Structure and complexity of a bacterial transcriptome. J. Bacteriol. 191, 3203–3211 (2009).
Sharma, C. M. et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255 (2010).
van Opijnen, T., Bodi, K. L. & Camilli, A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nature Methods 6, 767–772 (2009).
Langridge, G. C. et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19, 2308–2316 (2009).
Grainger, D. et al. Direct methods for studying transcription regulatory proteins and RNA polymerase in bacteria. Curr. Opin. Microbiol. 12, 531–535 (2009).
Branton, D. et al. The potential and challenges of nanopore sequencing. Nature Biotech. 26, 1146–1153 (2008).
Pallen, M. J. & Loman, N. J. Are diagnostic and public health bacteriology ready to become branches of genomic medicine? Genome Med. 3, 53 (2011).
Adey, A. et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Res. 11, R119 (2010).
Glenn, T. C. A field guide to next generation DNA sequencers. Mol. Ecol. Res. 11, 759–769 (2011).
Acknowledgements
The authors thank the anonymous reviewers for their help and suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Mark J. Pallen was a winner of an Ion Personal Genome Machine (PGM) (from Ion Torrent, part of Life Technologies) in the European Ion PGM Grant Programme. Nicholas J. Loman has received expenses to speak at an Ion Torrent meeting organized by Life Technologies and has received honoraria and expenses from Illumina to speak at Illumina meetings. Chrystala Constantinidou, Jacqueline Z. M. Chan, Mihail Halachev, Martin Sergeant, Charles W. Penn and Esther R. Robinson declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Available read mapping and de novo assembly software for next-generation sequence data (PDF 275 kb)
Related links
Rights and permissions
About this article
Cite this article
Loman, N., Constantinidou, C., Chan, J. et al. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10, 599–606 (2012). https://doi.org/10.1038/nrmicro2850
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2850
This article is cited by
-
A review on microplastic pollution in the mangrove wetlands and microbial strategies for its remediation
Environmental Science and Pollution Research (2022)
-
Development of a robust protocol for the characterization of the pulmonary microbiota
Communications Biology (2021)
-
An accessible, efficient and global approach for the large-scale sequencing of bacterial genomes
Genome Biology (2021)
-
Advances in monitoring soil microbial community dynamic and function
Journal of Applied Genetics (2020)