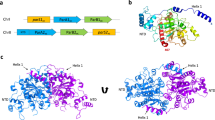
Key Points
-
ParMRC is an independent DNA segregation system that is carried by some low-copy-number plasmids and is sufficient to ensure that plasmids are not lost during host cell division.
-
ParMRC is composed of just three components: a filament-forming, actin-like ATPase motor (ParM), a DNA-binding adaptor (ParR) and a centromere-like DNA region (parC).
-
Plasmid segregation is driven by ParM filaments, which are dynamically unstable unless capped at both ends by a plasmid-bound ParR–parC complex. Once stabilized, ParM filaments move the attached plasmids in opposite directions, so driving the DNA apart.
-
ParM filaments assemble into small bundles in the cell, and these probably move clusters of plasmids apart. There are no known host cell factors required for segregation.
-
ParM is structurally related to eukaryotic actin and forms similar ATP-dependent double filaments, although the filaments wrap around one another with opposite handedness. Despite the structural similarity, the dynamics of ParM have three unique characteristics that differ from those of actin dynamics: equal bipolar elongation, dynamic instability and a high rate of spontaneous nucleation. These properties are tuned to enable ParM filaments to search the cell space for plasmid-bound ParR–parC complexes using a 'search and capture' mechanism.
-
The ParR–parC nucleoprotein cap forms a large solenoid structure that binds to the very tips of ParM filaments and prevents filament disassembly. The exact structure and molecular mechanism of this complex are unknown.
Abstract
The ParMRC plasmid partitioning apparatus is one of the best characterized systems for bacterial DNA segregation. Bundles of actin-like filaments are used to push plasmids to opposite poles of the cell, whereupon they are stably inherited on cell division. This plasmid-encoded system comprises just three components: an actin-like protein, ParM, a DNA-binding adaptor protein, ParR, and a centromere-like region, parC. The properties and interactions of these components have been finely tuned to enable ParM filaments to search the cell space for plasmids and then move ParR–parC-bound DNA molecules apart. In this Review, we look at some of the most exciting questions in the field concerning the exact molecular mechanisms by which the components of this self-contained system modulate one another's activity to achieve bipolar DNA segregation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ebersbach, G. & Gerdes, K. Plasmid segregation mechanisms. Annu. Rev. Genet. 39, 453–479 (2005).
Ghosh, S. K., Hajra, S., Paek, A. & Jayaram, M. Mechanisms for chromosome and plasmid segregation. Annu. Rev. Biochem. 75, 211–241 (2006).
Schumacher, M. A. Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 412, 1–18 (2008). A recent review describing the structural basis of the major plasmid segregation systems.
Gerdes, K., Howard, M. & Szardenings, F. Pushing and pulling in prokaryotic DNA segregation. Cell 141, 927–942 (2010).
Löwe, J. & Amos, L. A. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int. J. Biochem. Cell Biol. 41, 323–329 (2009). A description of nucleotide-driven dynamic filaments as 'cytomotive filaments', and a discussion of the evolutionary and mechanistic implications.
Ni, L., Xu, W., Kumaraswami, M. & Schumacher, M. A. Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc. Natl Acad. Sci. USA 107, 11763–11768 (2010).
Simpson, A. E., Skurray, R. A. & Firth, N. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J. Bacteriol. 185, 2143–2152 (2003).
Gerdes, K., Larsen, J. E. & Molin, S. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 161, 292–298 (1985).
Møller-Jensen, J. et al. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell 12, 1477–1487 (2003). The first immunofluorescence images showing labelled cellular ParM filaments and labelled plasmids. The filaments run along the length of the cell, with plasmids at both ends. This is also the first description of an insertional polymerization mechanism.
Garner, E. C., Campbell, C. S., Weibel, D. B. & Mullins, R. D. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315, 1270–1274 (2007). The entire ParMRC complex is reconstituted in vitro , and ParM filaments are seen to move ParR– parC -bound polystyrene beads apart. This is also the first description of the 'search and capture' mechanism.
Schumacher, M. A. et al. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature 450, 1268–1271 (2007). The co-crystal structure of pSK41-encoded ParR and short fragments of parC DNA, showing parC bound to the RHH 2 surface on the outside surface of the ParR helix.
Popp, D. et al. Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation. J. Biol. Chem. 285, 101300–101340 (2010).
Møller-Jensen, J., Ringgaard, S., Mercogliano, C. P., Gerdes, K. & Löwe, J. Structural analysis of the ParR/parC plasmid partition complex. EMBO J. 26, 4413–4422 (2007). The first crystal structure of ParR, in which ParR alone is crystallized in a continuous helical arrangement. This study includes an EM demonstration of ParR– parC rings and an analysis of DNA binding through mutagenesis of parR.
Derman, A. I. et al. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol. Microbiol. 73, 534–552 (2009).
Becker, E. et al. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 25, 5919–5931 (2006).
Polka, J. K., Kollman, J. M., Agard, D. A. & Mullins, R. D. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J. Bacteriol. 191, 6219–6230 (2009).
Popp, D. et al. Polymeric structures and dynamic properties of the bacterial actin AlfA. J. Mol. Biol. 397, 1031–1041 (2010).
Garner, E. C., Campbell, C. S. & Mullins, R. D. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306, 1021–1025 (2004). The first demonstration of dynamic instability and bipolar elongation for ParM, and a detailed analysis of the dynamic properties of ParM compared with those of actin.
Popp, D. et al. Concerning the dynamic instability of actin homolog ParM. Biochem. Biophys. Res. Commun. 353, 109–114 (2007).
Mitchison, T. & Kirschner, M. Dynamic instability of microtubule growth. Nature 312, 237–242 (1984).
Pfaendtner, J., Lyman, E., Pollard, T. D. & Voth, G. A. Structure and dynamics of the actin filament. J. Mol. Biol. 396, 252–263 (2010).
Bork, P., Sander, C. & Valencia, A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl Acad. Sci. USA 89, 7290–7294 (1992).
Jensen, R. B. & Gerdes, K. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J. Mol. Biol. 269, 505–513 (1997).
van den Ent, F., Møller-jensen, J., Amos, L. A., Gerdes, K. & Löwe, J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 21, 6935–6943 (2002). The first crystal structure of plasmid R1-encoded ParM, and the first molecular model of ParM double filaments.
Carballido-Lopez, R. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 70, 888–909 (2006).
Jones, L. J., Carballido-Lopez, R. & Errington, J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104, 913–922 (2001).
van den Ent, F., Amos, L. & Löwe, J. Bacterial ancestry of actin and tubulin. Curr. Opin. Microbiol. 4, 634–638 (2001).
van den Ent, F., Amos, L. A. & Löwe, J. Prokaryotic origin of the actin cytoskeleton. Nature 413, 39–44 (2001).
Komeili, A., Li, Z., Newman, D. K. & Jensen, G. J. Magnetosomes are cell membrane invaginations organised by the actin-like protein MamK. Science 311, 242–245 (2006).
Taoka, A., Asada, R., Wu, L. F. & Fukumori, Y. Polymerisation of the actin-like protein MamK, which is associated with magnetosomes. J. Bacteriol. 189, 8737–8740 (2007).
Popp, D. et al. Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 27, 570–579 (2008).
Orlova, A. et al. The structure of bacterial ParM filaments. Nature Struct. Mol. Biol. 14, 921–926 (2007).
Galkin, V. E., Orlova, A., Rivera, C., Mullins, R. D. & Egelman, E. H. Structural polymorphism of the ParM filament and dynamic instability. Structure 17, 1253–1264 (2009).
Popp, D. et al. Filament structure, organization and dynamics in MreB sheets. J. Biol. Chem. 21, 15858–15865 (2010).
Holmes, K., Popp, D., Gebhard, W. & Kabsch, W. Atomic model of the actin filament. Nature 347, 44–49 (1990).
Holmes, K. Actin in a twist. Nature 457, 389–390 (2009).
Oda, T., Iwasa, M., Aihara, T., Maeda, Y. & Narita, A. The nature of the globular- to fibrous-actin transition. Nature 457, 441–445 (2009).
Salje, J., Zuber, B. & Löwe, J. Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science 323, 509–512 (2009). A direct observation of ParM filaments in E. coli using cellular cryo-EM techniques.
Campbell, C. S. & Mullins, R. D. In vivo visualization of type II plasmid segregation: bacterial actin filaments pushing plasmids. J. Cell Biol. 179, 1059–1066 (2007). The first observation of dynamic ParM filaments and plasmids in live E. coli cells.
Popp, D., Narita, A., Iwasa, M., Maeda, Y. & Robinson, R. C. Molecular mechanism of bundle formation by the bacterial actin ParM. Biochem. Biophys. Res. Commun. 391, 1598–1603 (2010).
Dam, M. & Gerdes, K. Partitioning of plasmid R1. Ten direct repeats flanking the parA promoter constitute a centromere-like partition site parC, that expresses incompatibility. J. Mol. Biol. 236, 1289–1298 (1994).
Jensen, R. B., Dam, M. & Gerdes, K. Partitioning of plasmid R1. The parA operon is autoregulated by ParR and its transcription is highly stimulated by a downstream activating element. J. Mol. Biol. 236, 1299–1309 (1994).
Jensen, R. B., Lurz, R. & Gerdes, K. Mechanism of DNA segregation in prokaryotes: replicon pairing by parC of plasmid R1. Proc. Natl Acad. Sci. USA 95, 8550–8555 (1998).
Salje, J. & Löwe, J. Bacterial actin: architecture of the ParMRC plasmid DNA partitioning complex. EMBO J. 27, 2230–2238 (2008). A demonstration of the 1/1 stochiometry between ParM filaments and the ParR– parC complex, and a description of the clamp model for ParMRC.
Choi, C. L., Claridge, S. A., Garner, E. C., Alivisatos, A. P. & Mullins, R. D. Protein-nanocrystal conjugates support a single filament polymerization model in R1 plasmid segregation. J. Biol. Chem. 283, 28081–28086 (2008).
Davey, M. J. & Funnell, B. E. The P1 plasmid partition protein ParA. A role for ATP in site-specific DNA binding. J. Biol. Chem. 269, 29908–29913 (1994).
Davey, M. J. & Funnell, B. E. Modulation of the P1 plasmid partition protein ParA by ATP, ADP, and P1 ParB. J. Biol. Chem. 272, 15286–15292 (1997).
Davis, M. A., Martin, K. A. & Austin, S. J. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol. Microbiol. 6, 1141–1147 (1992).
Golovanov, A. P., Barilla, D., Golovanova, M., Hayes, F. & Lian, L. Y. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon–helix–helix structure. Mol. Microbiol. 50, 1141–1153 (2003).
Weitao, T., Dasgupta, S. & Nordström, K. Plasmid R1 is present as clusters in the cells of Escherichia coli. Plasmid 43, 200–204 (2000).
Møller-Jensen, J., Jensen, R. B., Löwe, J. & Gerdes, K. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21, 3119–3127 (2002). The first demonstration of dynamic filament formation by ParM, and the first molecular model for ParMRC plasmid segregation.
Nordström, K., Molin, S. & Aagaard-Hansen, H. Partitioning of plasmid R1 in Escherichia coli. I. Kinetics of loss of plasmid derivatives deleted of the par region. Plasmid 4, 215–227 (1980).
Gordon, S., Rech, J., Lane, D. & Wright, A. Kinetics of plasmid segregation in Escherichia coli. Mol. Microbiol. 51, 461–469 (2004).
Sengupta, M., Nielsen, H. J., Youngren, B. & Austin, S. P1 plasmid segregation: accurate redistribution by dynamic plasmid pairing and separation. J. Bacteriol. 192, 1175–1183 (2010).
Ringgaard, S., van Zon, J., Howard, M. & Gerdes, K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc. Natl Acad. Sci. USA 106, 19369–19374 (2009).
Tanaka, T. Functional analysis of the stability determinant AlfB of pBET131, a miniplasmid derivative of Bacillus subtilis (natto) plasmid pLS32. J. Bacteriol. 192, 1221–1230 (2010).
Greenfield, D. et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 7, e1000137 (2009).
Milne, J. L. & Subramaniam, S. Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nature Rev. Microbiol. 7, 666–675 (2009).
Li, Z. & Jensen, G. J. Electron cryotomography: a new view into microbial ultrastructure. Curr. Opin. Microbiol. 12, 333–340 (2009).
Raskin, D. M. & de Boer, P. A. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA 96, 4971–4976 (1999).
Cordell, S. C. & Löwe, J. Crystal structure of the bacterial cell division regulator MinD. FEBS Lett. 492, 160–165 (2001).
Michie, K. & Löwe, J. Dynamic filaments of the bacterial cytoskeleton. Annu. Rev. Biochem. 75, 467–492 (2006).
Lee, P. S. & Grossman, A. D. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 60, 853–869 (2006).
Murray, H. & Errington, J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135, 74–84 (2008).
Gruber, S. & Errington, J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137, 685–696 (2009).
Rodionov, O., Lobocka, M. & Yarmolinsky, M. Silencing of genes flanking the P1 plasmid centromere. Science 283, 546–549 (1999).
Surtees, J. A. & Funnell, B. E. The DNA binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 276, 12385–12394 (2001).
Rodionov, O. & Yarmolinsky, M. Plasmid partitioning and the spreading of P1 partition protein ParB. Mol. Microbiol. 52, 1215–1223 (2004).
Schumacher, M. A. & Funnell, B. E. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature 438, 516–519 (2005).
Schumacher, M. A., Mansoor, A. & Funnell, B. E. Structure of a four-way bridged ParB-DNA complex provides insight into P1 segrosome assembly. J. Biol. Chem. 282, 10456–10464 (2007).
Lim, G. E., Derman, A. I. & Pogliano, J. Bacterial DNA segregation by dynamic SopA polymers. Proc. Natl Acad. Sci. USA 102, 17658–17663 (2005).
Castaing, J. P., Bouet, J. Y. & Lane, D. F plasmid partition depends on interaction of SopA with non-specific DNA. Mol. Microbiol. 70, 1000–1011 (2008).
Bouet, J. Y., Ah-Seng, Y., Benmeradi, N. & Lane, D. Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol. Microbiol. 63, 468–481 (2007).
Barilla, D., Rosenberg, M. F., Nobbmann, U. & Hayes, F. Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO J. 24, 1453–1464 (2005).
Barilla, D., Carmelo, E. & Hayes, F. The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc. Natl Acad. Sci. USA 104, 1811–1816 (2007).
Dunham, T. D., Xu, W., Funnell, B. E. & Schumacher, M. A. Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA. EMBO J. 28, 1792–1802 (2009).
Ebersbach, G. et al. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol. Microbiol. 61, 1428–1442 (2006).
Hatano, T., Yamaichi, Y. & Niki, H. Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol. Microbiol. 64, 1198–1213 (2007).
Leonard, T. A., Møller-Jensen, J. & Lowe, J. Towards understanding the molecular basis of bacterial DNA segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 523–535 (2005).
Suefuji, K. V. R., RayChaudhuri, D. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids and MinE. Proc. Natl Acad. Sci. USA 99, 16776–16781 (2002).
Larsen, R. A. et al. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 21, 1340–1352 (2007).
Berry, C. et al. Complete sequence and organisation of pBtoxis, the toxin-encoding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 72, 5082–5095 (2002).
Tang, M., Bideshi, D. K., Park, H. W. & Federici, B. A. Minireplicon from pBtoxis of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 72, 6948–6954 (2006).
Tinsley, E. & Khan, S. A. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J. Bacteriol. 188, 2829–2835 (2006).
Robinett, C. C. et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135, 1685–1700 (1996).
Lemon, K. P. & Grossman, A. D. Movement of replicating DNA through a stationary replisome. Mol. Cell 6, 1321–1330 (2000).
Wang, X., Reyes-Lamothe, R. & Sherratt, D. J. Visualizing genetic loci and molecular machines in living bacteria. Biochem. Soc. Trans. 36, 749–753 (2008).
Vorobiev, S. et al. The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl Acad. Sci. USA 100, 5760–5765 (2003).
Galkin, V. E., VanLoock, M. S., Orlova, A. & Egelman, E. H. A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin's sequence and structure. Curr. Biol. 12, 570–575 (2002).
Acknowledgements
The authors are grateful to K. Gerdes for helpful comments on the manuscript and to J. Møller-Jensen for ongoing collaborations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Walker A protein
-
Protein that contains a Walker A motif (GXXXGKT; where X is any amino acid), and is involved in the nucleotide binding of many ATP-requiring enzymes.
- Tubulin
-
Basic subunit of microtubules. Tubulin comes in two forms, α-tubulin and β-tubulin, which form heterodimers that make up microtubules.
- Filamentous actin
-
Flexible, helical polymer of G-actin monomers that is 5–9nm in diameter. This polymer is polar, displaying a plus end and a minus end.
- Microtubule
-
Hollow tube, 25nm in diameter, that is formed by the lateral association of 13 protofilaments, which are themselves polymers of α-tubulin and β-tubulin subunits.
- Cofilin
-
Actin-binding protein that promotes disassembly at the minus ends of actin filaments.
- Barbed end
-
The plus end of the polar F-actin polymer, which is more active than the minus end with regard to the incorporation of G-actin into the polymer.
- Magnetosome
-
Unique intracellular structure that is found in magnetotactic bacteria and comprises a magnetic mineral crystal surrounded by a lipid bilayer membrane.
- Nucleoid
-
Distinct region in the bacterial cytoplasm that harbours the chromosomal DNA.
- Formin
-
Protein that contains a formin homology 2 (FH2) domain and promotes actin assembly. Formin binds to the ends of actin filaments.
Rights and permissions
About this article
Cite this article
Salje, J., Gayathri, P. & Löwe, J. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol 8, 683–692 (2010). https://doi.org/10.1038/nrmicro2425
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2425
This article is cited by
-
World War II, Sex and Antibiotics
Resonance (2023)
-
World War II, Sex and Antibiotics - II
Resonance (2023)
-
Towards a better understanding of antimicrobial resistance dissemination: what can be learnt from studying model conjugative plasmids?
Military Medical Research (2022)
-
Revealing biophysical properties of KfrA-type proteins as a novel class of cytoskeletal, coiled-coil plasmid-encoded proteins
BMC Microbiology (2021)
-
Putative plasmid prophages of Bacillus cereus sensu lato may hold the key to undiscovered phage diversity
Scientific Reports (2021)