Key Points
-
Ribonuclease P (RNase P) is a ribonucleoprotein particle that catalyses maturation of the 5′ end of transfer RNA (tRNA) by cleavage of precursor-specific sequences. RNase P is found in cells from all three domains of life: the Bacteria, Eukarya and Archaea. It contains a highly conserved catalytic RNA component — a unique, natural ribozyme that conducts multiple turnovers with a broad substrate specificity.
-
RNase P RNA is a metalloenzyme that catalyses hydrolysis of a specific internucleotide phosphodiester bond through an SN2-like nucleophilic substitution mechanism. Divalent metal ions participate directly in the reaction through a variation of a two-metal-ion mechanism that is typical of the large ribozymes such as group-I introns and many protein-based phosphoryl-transfer enzymes.
-
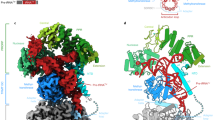
Early perspectives into the structure of the RNase P RNA came from phylogenetic comparative studies, complemented by photo-affinity crosslinking and computer modelling. Recent crystallographic studies of bacterial RNase P RNA corroborate the results of early structural, biochemical and biophysical studies, and provide a new and detailed view of the overall structural organization of the ribozyme, its substrate-binding interface and the proposed chemically active site.
-
Based on the newly available structural information, the functional complexes of the bacterial RNase P RNA with its protein cofactor and with the product tRNA have been modelled. Considering the relatively low resolution of the available crystal structures, many details of the RNA interaction with the protein component and with the substrate are still uncertain, and the atomic details of how the RNA participates in catalysis remain to be elucidated.
Abstract
Ribonuclease P (RNase P) is a ubiquitous endonuclease that catalyses the maturation of the 5′ end of transfer RNA (tRNA). Although it carries out a biochemically simple reaction, RNase P is a complex ribonucleoprotein particle composed of a single large RNA and at least one protein component. In bacteria and some archaea, the RNA component of RNase P can catalyse tRNA maturation in vitro in the absence of proteins. The discovery of the catalytic activity of the bacterial RNase P RNA triggered numerous mechanistic and biochemical studies of the reactions catalysed by the RNA alone and by the holoenzyme and, in recent years, structures of individual components of the RNase P holoenzyme have been determined. The goal of the present review is to summarize what is known about the bacterial RNase P, and to bring together the recent structural results with extensive earlier biochemical and phylogenetic findings.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Frank, D. N. & Pace, N. R. Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67, 153–180 (1998).
Walker, S. C. & Engelke, D. R. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 41, 77–102 (2006).
Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857 (1983). Reports the discovery that the RNA of bacterial RNase P is catalytically proficient in the absence of the protein moiety of the enzyme.
Pannucci, J. A., Haas, E. S., Hall, T. A., Harris, J. K. & Brown, J. W. RNase P RNAs from some archaea are catalytically active. Proc. Natl Acad. Sci. USA 96, 7803–7808 (1999).
Schedl, P. & Primakoff, P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc. Natl Acad. Sci. USA 70, 2091–2095 (1973).
Apirion, D. Genetic mapping and some characterization of the rnpA49 mutation of Escherichia coli that affects the RNA-processing enzyme ribonuclease P. Genetics 94, 291–299 (1980).
Waugh, D. S. & Pace, N. R. Complementation of an RNase P RNA (rnpB) gene deletion in Escherichia coli by homologous genes from distantly related eubacteria. J. Bacteriol. 172, 6316–6322 (1990).
Chamberlain, J. R., Lee, Y., Lane, W. S. & Engelke, D. R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 12, 1678–1690 (1998). The most thorough of the studies of the eukaryal RNase P.
Hartmann, E. & Hartmann, R. K. The enigma of ribonuclease P evolution. Trends Genet. 19, 561–569 (2003).
Jarrous, N. Human ribonuclease P: subunits, function, and intranuclear localization. RNA 8, 1–7 (2002).
Evans, D., Marquez, S. M. & Pace, N. R. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 31, 333–341 (2006).
Kirsebom, L. A. RNase P RNA-mediated catalysis. Biochem. Soc. Trans. 30, 1153–1158 (2002).
Harris, M. E. & Christian, E. L. Recent insights into the structure and function of the ribonucleoprotein enzyme ribonuclease P. Curr. Opin. Struct. Biol. 13, 325–333 (2003).
Torres-Larios, A., Swinger, K. K., Pan, T. & Mondragon, A. Structure of ribonuclease P — a universal ribozyme. Curr. Opin. Struct. Biol. 16, 327–335 (2006).
Peck-Miller, K. A. & Altman, S. Kinetics of the processing of the precursor to 4.5S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J. Mol. Biol. 221, 1–5 (1991).
Bothwell, A. L. M., Stark, B. C. & Altman, S. Ribonuclease P substrate specificity: cleavage of a bacteriophage ø80-induced RNA. Proc. Natl Acad. Sci. USA 73, 1912–1916 (1976).
Guerrier-Takada, C., Van Belkum, A., Pleij, C. W. & Altman, S. Novel reactions of RNase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell 53, 267–272 (1988).
Komine, Y., Kitabatake, M., Yokogawa, T., Nishikawa, K. & Inokuchi, H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl Acad. Sci. USA 91, 9223–9227 (1994).
Alifano, P. et al. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 8, 3021–3031 (1994).
Hartmann, R. K., Heinrich, J., Schlegl, J. & Schuster, H. Precursor of C4 antisense RNA of bacteriophages P1 and P7 is a substrate for RNase P of Escherichia coli. Proc. Natl Acad. Sci. USA 92, 5822–5826 (1995).
Altman, S., Wesolowski, D., Guerrier-Takada, C. & Li, Y. RNase P cleaves transient structures in some riboswitches. Proc. Natl Acad. Sci. USA (2005).
Altman, S. & Smith, J. D. Tyrosine tRNA precursor molecule polynucleotide sequence. Nature New Biol. 233, 35–39 (1971). Reports the discovery of the processing enzyme RNase P. The 'P' stands for 'processing'.
Smith, D. & Pace, N. R. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry 32, 5273–5281 (1993).
Beebe, J. A. & Fierke, C. A. A kinetic mechanism for cleavage of precursor tRNAAsp catalyzed by the RNA component of Bacillus subtilis ribonuclease P. Biochemistry 33, 10294–10304 (1994).
Persson, T., Cuzic, S. & Hartmann, R. K. Catalysis by RNase P RNA: unique features and unprecedented active site plasticity. J. Biol. Chem. 278, 43394–43401 (2003).
Cassano, A. G., Anderson, V. E. & Harris, M. E. Analysis of solvent nucleophile isotope effects: evidence for concerted mechanisms and nucleophilic activation by metal coordination in nonenzymatic and ribozyme-catalyzed phosphodiester hydrolysis. Biochemistry 43, 10547–10559 (2004).
Smith, D., Burgin, A. B., Haas, E. S. & Pace, N. R. Influence of metal ions on the ribonuclease P reaction. Distinguishing substrate binding from catalysis. J. Biol. Chem. 267, 2429–2436 (1992).
Brannvall, M. & Kirsebom, L. A. Metal ion cooperativity in ribozyme cleavage of RNA. Proc. Natl Acad. Sci. USA 98, 12943–12947 (2001).
Cuzic, S. & Hartmann, R. K. Studies on Escherichia coli RNase P RNA with Zn2+ as the catalytic cofactor. Nucleic Acids Res. 33, 2464–2474 (2005).
Kikovska, E., Mikkelsen, N. E. & Kirsebom, L. A. The naturally trans-acting ribozyme RNase P RNA has leadzyme properties. Nucleic Acids Res. 33, 6920–6930 (2005).
Beebe, J. A., Kurz, J. C. & Fierke, C. A. Magnesium ions are required by Bacillus subtilis ribonuclease P RNA for both binding and cleaving precursor tRNAAsp. Biochemistry 35, 10493–10505 (1996).
Pan, T. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry 34, 902–909 (1995). Shows that the RNase P RNA structure consists of independently folding domains.
Fang, X. W., Pan, T. & Sosnick, T. R. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nature Struct. Biol. 6, 1091–1095 (1999).
Kent, O., Chaulk, S. G. & Macmillan, A. M. Kinetic analysis of the M1 RNA folding pathway. J. Mol. Biol. 304, 699–705 (2000).
Warnecke, J. M., Furste, J. P., Hardt, W. D., Erdmann, V. A. & Hartmann, R. K. Ribonuclease P (RNase P) RNA is converted to a Cd2+-ribozyme by a single Rp-phosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc. Natl Acad. Sci. USA 93, 8924–8928 (1996).
Warnecke, J. M., Held, R., Busch, S. & Hartmann, R. K. Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis. J. Mol. Biol. 290, 433–445 (1999).
Steitz, T. A. & Steitz, J. A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 (1993). A general model for two-metal-promoted trans esterification involving the phosphodiester bond.
Yang, W., Lee, J. Y. & Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006).
Stahley, M. R. & Strobel, S. A. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science 309, 1587–1590 (2005).
Brautigam, C. A. & Steitz, T. A. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 8, 54–63 (1998).
Jager, J. & Pata, J. D. Getting a grip: polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 9, 21–28 (1999).
Nowotny, M., Gaidamakov, S. A., Crouch, R. J. & Yang, W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016 (2005).
Pingoud, A., Fuxreiter, M., Pingoud, V. & Wende, W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 62, 685–707 (2005).
Stoddard, B. L. Homing endonuclease structure and function. Q. Rev. Biophys. 38, 49–95 (2005).
Fox, G. W. & Woese, C. R. 5S RNA secondary structure. Nature 256, 505–507 (1975).
Pace, N. R., Smith, D. K., Olsen, G. J. & James, B. D. Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA — a review. Gene 82, 65–75 (1989).
Brown, J. W. et al. Comparative analysis of ribonuclease P RNA using gene sequences from natural microbial populations reveals tertiary structural elements. Proc. Natl Acad. Sci. USA 93, 3001–3006 (1996). First sequence-comparative RNA-structure analysis based on genes from complex natural microbial communities.
Pace, N. R. & Brown, J. W. Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J. Bacteriol. 177, 1919–1928 (1995).
Haas, E. S. & Brown, J. W. Evolutionary variation in bacterial RNase P RNAs. Nucleic Acids Res. 26, 4093–4099 (1998).
Siegel, R. W., Banta, A. B., Haas, E. S., Brown, J. W. & Pace, N. R. Mycoplasma fermentans simplifies our view of the catalytic core of ribonuclease P RNA. RNA 2, 452–462 (1996).
Harris, M. E. et al. Use of photoaffinity crosslinking and molecular modeling to analyze the global architecture of ribonuclease P RNA. EMBO J. 13, 3953–3963 (1994).
Westhof, E. & Altman, S. Three-dimensional working model of M1 RNA, the catalytic RNA subunit of ribonuclease P from Escherichia coli. Proc. Natl Acad. Sci. USA 91, 5133–5137 (1994).
Harris, M. E., Kazantsev, A. V., Chen, J. L. & Pace, N. R. Analysis of the tertiary structure of the ribonuclease P ribozyme–substrate complex by site-specific photoaffinity crosslinking. RNA 3, 561–576 (1997).
Thomas, B. C., Kazantsev, A. V., Chen, J. L. & Pace, N. R. Photoaffinity cross-linking and RNA structure analysis. Methods Enzymol. 318, 136–147 (2000). Methodological review of the photo-affinity crosslinking agents and modelling strategies used to determine large-scale RNA structure.
Chen, J. L., Nolan, J. M., Harris, M. E. & Pace, N. R. Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. EMBO J. 17, 1515–1525 (1998).
Massire, C., Jaeger, L. & Westhof, E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 279, 773–793 (1998).
Chen, J. L. & Pace, N. R. Identification of the universally conserved core of ribonuclease P RNA. RNA 3, 557–560 (1997).
Harris, J. K., Haas, E. S., Williams, D., Frank, D. N. & Brown, J. W. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA 7, 220–232 (2001).
Marquez, S. M. et al. Structural implications of novel diversity in eucaryal RNase P RNA. RNA 11, 739–751 (2005).
Pfeiffer, T. et al. Effects of phosphorothioate modifications on precursor tRNA processing by eukaryotic RNase P enzymes. J. Mol. Biol. 298, 559–565 (2000).
Thomas, B. C., Chamberlain, J., Engelke, D. R. & Gegenheimer, P. Evidence for an RNA-based catalytic mechanism in eukaryotic nuclear ribonuclease P. RNA 6, 554–562 (2000).
Thomas, B. C., Li, X. & Gegenheimer, P. Chloroplast ribonuclease P does not utilize the ribozyme-type pre-tRNA cleavage mechanism. RNA 6, 545–553 (2000).
Torres-Larios, A., Swinger, K. K., Krasilnikov, A. S., Pan, T. & Mondragon, A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature 437, 584–587 (2005). A 3.8-Å X-ray crystal structure of an A-type RNase P RNA, that of Thermotoga maritima .
Kazantsev, A. V. et al. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl Acad. Sci. USA 102, 13992–13997 (2005). A 3.3-Å X-ray crystal structure of a B-type RNase P RNA, that of Bacillus stearothermophilus.
Loria, A. & Pan, T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA 2, 551–563 (1996).
Krasilnikov, A. S., Xiao, Y., Pan, T. & Mondragon, A. Basis for structural diversity in homologous RNAs. Science 306, 104–107 (2004).
Krasilnikov, A. S., Yang, X., Pan, T. & Mondragon, A. Crystal structure of the specificity domain of ribonuclease P. Nature 421, 760–764 (2003).
Fang, X. W. et al. The Bacillus subtilis RNase P holoenzyme contains two RNase P RNA and two RNase P protein subunits. RNA 7, 233–241 (2001).
Buck, A. H., Dalby, A. B., Poole, A. W., Kazantsev, A. V. & Pace, N. R. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 24, 3360–3368 (2005).
Moore, P. B. Structural motifs in RNA. Annu. Rev. Biochem. 68, 287–300 (1999).
Leontis, N. B. & Westhof, E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 13, 300–308 (2003).
Holbrook, S. R. RNA structure: the long and the short of it. Curr. Opin. Struct. Biol. 15, 302–308 (2005).
Cate, J. H. et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685 (1996).
Tinoco, I., Jr. RNA enzymes: putting together a large ribozyme. Curr. Biol. 6, 1374–1376 (1996).
Tamura, M. & Holbrook, S. R. Sequence and structural conservation in RNA ribose zippers. J. Mol. Biol. 320, 455–474 (2002).
Nissen, P., Ippolito, J. A., Ban, N., Moore, P. B. & Steitz, T. A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA 98, 4899–4903 (2001).
Cate, J. H. et al. RNA tertiary structure mediation by adenosine platforms. Science 273, 1696–1699 (1996).
Klosterman, P. S., Hendrix, D. K., Tamura, M., Holbrook, S. R. & Brenner, S. E. Three-dimensional motifs from the SCOR, structural classification of RNA database: extruded strands, base triples, tetraloops and U-turns. Nucleic Acids Res. 32, 2342–2352 (2004).
Krasilnikov, A. S. & Mondragon, A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA 9, 640–643 (2003).
Hardt, W. D., Warnecke, J. M., Erdmann, V. A. & Hartmann, R. K. Rp-phosphorothioate modifications in RNase P RNA that interfere with tRNA binding. EMBO J. 14, 2935–2944 (1995).
Hardt, W. D., Erdmann, V. A. & Hartmann, R. K. Rp-deoxy-phosphorothioate modification interference experiments identify 2′-OH groups in RNase P RNA that are crucial to tRNA binding. RNA 2, 1189–1198 (1996).
Heide, C., Pfeiffer, T., Nolan, J. M. & Hartmann, R. K. Guanosine 2-NH2 groups of Escherichia coli RNase P RNA involved in intramolecular tertiary contacts and direct interactions with tRNA. RNA 5, 102–116 (1999).
Siew, D., Zahler, N. H., Cassano, A. G., Strobel, S. A. & Harris, M. E. Identification of adenosine functional groups involved in substrate binding by the ribonuclease P ribozyme. Biochemistry 38, 1873–1883 (1999).
Heide, C., Busch, S., Feltens, R. & Hartmann, R. K. Distinct modes of mature and precursor tRNA binding to Escherichia coli RNase P RNA revealed by NAIM analyses. RNA 7, 553–564 (2001).
Heide, C., Feltens, R. & Hartmann, R. K. Purine N7 groups that are crucial to the interaction of Escherichia coli RNase P RNA with tRNA. RNA 7, 958–968 (2001).
Lagrandeur, T. E., Huttenhofer, A., Noller, H. F. & Pace, N. R. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 13, 3945–3952 (1994).
Odell, L., Huang, V., Jakacka, M. & Pan, T. Interaction of structural modules in substrate binding by the ribozyme from Bacillus subtilis RNase P. Nucleic Acids Res. 26, 3717–3723 (1998).
Burgin, A. B. & Pace, N. R. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 9, 4111–4118 (1990).
Harris, M. E. & Pace, N. R. Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA 1, 210–218 (1995).
Frank, D. N. & Pace, N. R. In vitro selection for altered divalent metal specificity in the RNase P RNA. Proc. Natl Acad. Sci. USA 94, 14355–14360 (1997).
Christian, E. L., Kaye, N. M. & Harris, M. E. Helix P4 is a divalent metal ion binding site in the conserved core of the ribonuclease P ribozyme. RNA 6, 511–519 (2000).
Schmitz, M. & Tinoco, I., Jr. Solution structure and metal-ion binding of the P4 element from bacterial RNase P RNA. RNA 6, 1212–1225 (2000).
Christian, E. L., Kaye, N. M. & Harris, M. E. Evidence for a polynuclear metal ion binding site in the catalytic domain of ribonuclease P RNA. EMBO J. 21, 2253–2262 (2002).
Crary, S. M., Kurz, J. C. & Fierke, C. A. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis ribonuclease P. RNA 8, 933–947 (2002).
Stams, T., Niranjanakumari, S., Fierke, C. A. & Christianson, D. W. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science 280, 752–755 (1998).
Kazantsev, A. V. et al. High-resolution structure of RNase P protein from Thermotoga maritima. Proc. Natl Acad. Sci. USA 100, 7497–7502 (2003).
Spitzfaden, C. et al. The structure of ribonuclease P protein from Staphylococcus aureus reveals a unique binding site for single-stranded RNA. J. Mol. Biol. 295, 105–115 (2000).
Wegscheid, B., Condon, C. & Hartmann, R. K. Type A and B RNase P RNAs are interchangeable in vivo despite substantial biophysical differences. EMBO Rep. 7, 411–417 (2006).
Gopalan, V., Baxevanis, A. D., Landsman, D. & Altman, S. Analysis of the functional role of conserved residues in the protein subunit of ribonuclease P from Escherichia coli. J. Mol. Biol. 267, 818–829 (1997).
Gopalan, V. et al. Mapping RNA–protein interactions in ribonuclease P from Escherichia coli using electron paramagnetic resonance spectroscopy. Biochemistry 38, 1705–1714 (1999).
Biswas, R., Ledman, D. W., Fox, R. O., Altman, S. & Gopalan, V. Mapping RNA–protein interactions in ribonuclease P from Escherichia coli using disulfide-linked EDTA–Fe. J. Mol. Biol. 296, 19–31 (2000).
Tsai, H. Y., Masquida, B., Biswas, R., Westhof, E. & Gopalan, V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol. 325, 661–675 (2003).
Niranjanakumari, S., Stams, T., Crary, S. M., Christianson, D. W. & Fierke, C. A. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl Acad. Sci. USA 95, 15212–15217 (1998).
Westhof, E., Wesolowski, D. & Altman, S. Mapping in three dimensions of regions in a catalytic RNA protected from attack by an Fe(II)–EDTA reagent. J. Mol. Biol. 258, 600–613 (1996).
Loria, A., Niranjanakumari, S., Fierke, C. A. & Pan, T. Recognition of a pre-tRNA substrate by the Bacillus subtilis RNase P holoenzyme. Biochemistry 37, 15466–15473 (1998).
Sharkady, S. M. & Nolan, J. M. Bacterial ribonuclease P holoenzyme crosslinking analysis reveals protein interaction sites on the RNA subunit. Nucleic Acids Res. 29, 3848–3856 (2001).
Rox, C., Feltens, R., Pfeiffer, T. & Hartmann, R. K. Potential contact sites between the protein and RNA subunit in the Bacillus subtilis RNase P holoenzyme. J. Mol. Biol. 315, 551–560 (2002).
Buck, A. H., Kazantsev, A. V., Dalby, A. B. & Pace, N. R. Structural perspective on the activation of RNase P RNA by protein. Nature Struct. Mol. Biol. 12, 958–964 (2005). Describes an in-gel footprinting technique for distinguishing native versus denatured RNase P complexes.
McClain, W. H., Guerrier-Takada, C. & Altman, S. Model substrates for an RNA enzyme. Science 238, 527–530 (1987). Reports the first use of 'designed' substrates for RNase P RNA.
Svard, S. G. & Kirsebom, L. A. Several regions of a tRNA precursor determine the Escherichia coli RNase P cleavage site. J. Mol. Biol. 227, 1019–1031 (1992).
Yuan, Y. & Altman, S. Substrate recognition by human RNase P: identification of small, model substrates for the enzyme. EMBO J. 14, 159–168 (1995).
Pan, T., Loria, A. & Zhong, K. Probing of tertiary interactions in RNA: 2′-hydroxyl–base contacts between the RNase P RNA and pre-tRNA. Proc. Natl Acad. Sci. USA 92, 12510–12514 (1995).
Loria, A. & Pan, T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry 36, 6317–6325 (1997).
Kirsebom, L. A. & Svard, S. G. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 13, 4870–4876 (1994).
Oh, B. K. & Pace, N. R. Interaction of the 3′-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 22, 4087–4094 (1994).
Hardt, W. D., Schlegl, J., Erdmann, V. A. & Hartmann, R. K. Kinetics and thermodynamics of the RNase P RNA cleavage reaction: analysis of tRNA 3′-end variants. J. Mol. Biol. 247, 161–172 (1995).
Pellegrini, O., Nezzar, J., Marchfelder, A., Putzer, H. & Condon, C. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis. EMBO J. 22, 4534–4543 (2003).
Nakanishi, K. & Nureki, O. Recent progress of structural biology of tRNA processing and modification. Mol. Cells 19, 157–166 (2005).
Vogel, A., Schilling, O., Spath, B. & Marchfelder, A. The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties. Biol. Chem. 386, 1253–1264 (2005).
Svard, S. G., Kagardt, U. & Kirsebom, L. A. Phylogenetic comparative mutational analysis of the base-pairing between RNase P RNA and its substrate. RNA 2, 463–472 (1996).
Busch, S., Kirsebom, L. A., Notbohm, H. & Hartmann, R. K. Differential role of the intermolecular base-pairs G292–C75 and G293–C74 in the reaction catalyzed by Escherichia coli RNase P RNA. J. Mol. Biol. 299, 941–951 (2000).
Brannvall, M., Pettersson, B. M. & Kirsebom, L. A. Importance of the +73/294 interaction in Escherichia coli RNase P RNA substrate complexes for cleavage and metal ion coordination. J. Mol. Biol. 325, 697–709 (2003).
Brannvall, M. & Kirsebom, L. A. Complexity in orchestration of chemical groups near different cleavage sites in RNase P RNA mediated cleavage. J. Mol. Biol. (2005).
Brannvall, M., Kikovska, E. & Kirsebom, L. A. Cross talk between the +73/294 interaction and the cleavage site in RNase P RNA mediated cleavage. Nucleic Acids Res. 32, 5418–5429 (2004).
Zahler, N. H., Christian, E. L. & Harris, M. E. Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA 9, 734–745 (2003).
Zahler, N. H., Sun, L., Christian, E. L. & Harris, M. E. The pre-tRNA nucleotide base and 2′-hydroxyl at N–1 contribute to fidelity in tRNA processing by RNase P. J. Mol. Biol. 345, 969–985 (2005).
Brannvall, M., Mattsson, J. G., Svard, S. G. & Kirsebom, L. A. RNase P RNA structure and cleavage reflect the primary structure of tRNA genes. J. Mol. Biol. 283, 771–783 (1998).
Loria, A. & Pan, T. Recognition of the 5′ leader and the acceptor stem of a pre-tRNA substrate by the ribozyme from Bacillus subtilis RNase P. Biochemistry 37, 10126–10133 (1998).
Hansen, A. et al. Exploring the minimal substrate requirements for trans-cleavage by RNase P holoenzymes from Escherichia coli and Bacillus subtilis. Mol. Microbiol. 41, 131–143 (2001).
Rueda, D., Hsieh, J., Day-Storms, J. J., Fierke, C. A. & Walter, N. G. The 5′ leader of precursor tRNAAsp bound to the Bacillus subtilis RNase P holoenzyme has an extended conformation. Biochemistry 44, 16130–16139 (2005).
Christian, E. L., Smith, K. M., Perera, N. & Harris, M. E. The P4 metal binding site in RNase P RNA affects active site metal affinity through substrate positioning. RNA 12, 1463–1467 (2006).
Yarus, M. How many catalytic RNAs? Ions and the Cheshire Cat conjecture. FASEB 7, 31–39 (1993).
Acknowledgements
The authors' research is supported by a grant from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Ribozyme
-
An enzyme that has an RNA as the catalytic component. RNase P belongs to the large ribozyme class, which also includes the self-splicing group-I and group-II introns.
- 4.5S RNA
-
In bacteria, the signal-recognition particle comprises 4.5S RNA and the Ffh protein.
- Transfer messenger RNA
-
(Tm RNA). Also known as SsrA. Involved in a trans-translation process that adds a C-terminal peptide tag to unfinished proteins at stalled ribosomes that targets the unfinished proteins for proteolysis.
- Riboswitch
-
A regulatory structure in an mRNA molecule that undergoes a conformational change induced by the binding of a small metabolite, and which results in activation or inactivation of the mRNA.
- SN2 nucleophilic reaction
-
A bimolecular nucleophilic substitution reaction that involves displacement of a leaving group by an attacking nucleophile. An SN2 reaction that involves phosphate esters (for example, an internucleotide phosphodiester) proceeds through the formation of a trigonal bipyramidal transition state.
- Quench-flow technique
-
A technique that allows the analysis of chemical reactions at the millisecond time scale.
- Hill analysis
-
A common technique that analyses cooperative binding of ligands to biomacromolecules (proteins and nucleic acids). Either equilibrium binding (for example, extent of saturation) or the rate of the ligand-dependent enzymatic reaction is followed as a function of the concentration of the ligand. Mathematical analysis of the binding function yields the number of cooperatively bound ligands as well as the affinity constant.
- Phosphorothioate substitution
-
A common chemical modification that substitutes a sulphur atom for one of the non-bridging oxygens in the phosphodiester linkage.
- Thiophilic
-
Having a high affinity for sulphur.
- Kinetic isotope-effect study
-
A kinetic isotope effect is a change in the rate of a chemical reaction owing to a substitution of a participating chemical group with an analogue that contains a different isotope of one of the constituent atoms. As kinetic isotope effects reflect changes in the vibration energy of the transition state of the reaction caused by substitution, they are a useful tool for studying the mechanism of chemical reactions.
- RNase H
-
An endoribonuclease that specifically hydrolyses the phosphodiester bonds of RNA that are hybridized to DNA. Does not digest single-stranded or double-stranded DNA.
- Homing endonucleases
-
A large class of endonuclease that recognize a specific sequence that flanks the homing endonuclease-encoding gene, but only when the homing endonuclease-encoding gene itself does not interrupt this sequence.
- A-minor interaction
-
A common long-range interaction in structured RNA molecules achieved by docking of a bulged nucleotide (usually adenine) into a minor groove of a double-stranded helical structure. Four types of A-minor interactions are known; not all are specific for adenine. A-minor interactions are considered to be a subclass of ribose-zipper interactions.
- Tetraloop
-
A four-base loop that caps a hairpin helix. Tetraloops often dock into other structural elements of RNA to provide long-range structural stability.
- Ribose zipper
-
An element of RNA structure that is characterized by consecutive hydrogen-bonding interactions between ribose 2′-hydroxyls from different regions of an RNA chain or between RNA chains.
- Dinucleotide platform
-
A structural feature in which folding of the RNA chain results in an in-plane, side-by-side positioning of consecutive bases.
- Nucleotide analogue interference mapping
-
(NAIM). A technique that involves random, but sequence-specific, incorporation of chemical modifications into a pool of RNA molecules (usually achieved by in vitro transcription in the presence of nucleotide analogues) that is followed by a selection for a certain property (for example, folding, catalytic activity or ligand binding). Biochemical analysis of the distribution of such modifications reveals sites in RNA where the chemical modification interferes with the RNA property of interest.
- S5 superfamily
-
A protein structural family that includes proteins and protein domains with a similar α–β-sandwich-like fold. The family is named after one of its members, a small subunit ribosomal protein S5.
Rights and permissions
About this article
Cite this article
Kazantsev, A., Pace, N. Bacterial RNase P: a new view of an ancient enzyme. Nat Rev Microbiol 4, 729–740 (2006). https://doi.org/10.1038/nrmicro1491
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1491
This article is cited by
-
An extended catalogue of ncRNAs in Streptomyces coelicolor reporting abundant tmRNA, RNase-P RNA and RNA fragments derived from pre-ribosomal RNA leader sequences
Archives of Microbiology (2022)
-
Genome-wide discovery of structured noncoding RNAs in bacteria
BMC Microbiology (2019)
-
The unique tRNASec and its role in selenocysteine biosynthesis
Amino Acids (2018)
-
NMR resonance assignments of RNase P protein from Thermotoga maritima
Biomolecular NMR Assignments (2018)
-
Conditionally disordered proteins: bringing the environment back into the fold
Cellular and Molecular Life Sciences (2017)