Key Points
-
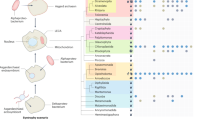
Viruses that infect the Crenarchaeota are represented by 20 cultured isolates that can infect members of four genera. These viruses represent up to nine new families of viruses owing to their distinctive morphologies and unique genome contents.
-
Acidic hot springs, with temperatures greater than 75°C and pH lower than 4.0, are typically dominated by a wide diversity of crenarchaea. Although molecular signatures of crenarchaea have been detected in a wide range of environments, all of the cultured members are thermophilic and many are acidophilic.
-
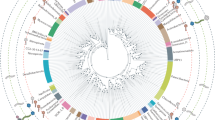
Virus morphology might indicate evolutionary history. The spindle-shaped morphology seems to be limited to the Archaea with isolates infecting both the Euryarchaeota and the Crenarchaeota. The icosahedral morphology seems to share an evolutionary connection with viruses infecting the Euryarchaeota as well as the Bacteria and Eukarya.
-
All of the isolated crenarchaeal viruses contain dsDNA genomes, most of which have been completely sequenced. Sequencing has revealed few ORFs with similarity to known genes in the public databases, however, threading algorithms and structural studies have led to possible functions being proposed for some of the proteins. As more genomes are sequenced it is becoming clear that some of the genes are shared between individual viruses and groups of viruses.
-
Little is known about the replication cycle for most of the isolated crenarcheal viruses. SSV-like viruses are known to integrate into the genome of the host, as does ATV. Unlike the SSV-like viruses, ATV lyses the host cell when it is induced and then undergoes an extracellular maturation under conditions ideal for its host.
-
Viruses that infect the Crenarchaeota can be isolated from around the world. Although viruses with similar morphology can be isolated from geographically separated hot springs, it is unclear if the viruses are genetically distinct or globally linked. The extent and distribution of the total diversity of crenarchaeal viruses remains to be determined.
Abstract
The discovery of archaeal viruses provides insights into the fundamental biochemistry and evolution of the Archaea. Recent studies have identified a wide diversity of archaeal viruses within the hot springs of Yellowstone National Park and other high-temperature environments worldwide. These viruses are often morphologically unique and code for genes with little similarity to other known genes in the biosphere, a characteristic that has complicated efforts to trace their evolutionary history. Comparative genomics combined with structural analysis indicate that spindle-shaped virus lineages might be unique to the Archaea, whereas other icosahedral viruses might share a common lineage with viruses of Bacteria and Eukarya. These studies provide insights into the evolutionary history of viruses in all three domains of life.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Prangishvili, D. & Garrett, R. A. Viruses of hyperthermophilic Crenarchaea. Trends Microbiol. 13, 535–542 (2005). This recent review provides more in-depth coverage of the structure of crenarchaeal virus particles and their genome, including replication and transcription.
Arnold, H. P., Ziese, U. & Zillig, W. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology 272, 409–416 (2000).
Häring, M., Rachel, R., Peng, X., Garrett, R. A. & Prangishvili, D. Viral diversity in hot springs of Pozzuoli, Italy, and characterization of a unique archaeal virus, Acidianus bottle-shaped virus, from a new family, the Ampullaviridae. J. Virol. 79, 9904–9911 (2005).
Bettstetter, M., Peng, X., Garrett, R. A. & Prangishvili, D. AFV1, a novel virus infecting hyperthermophilic archaea of the genus Acidianus. Virology 315, 68–79 (2003).
Vestergaard, G. et al. A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology 336, 83–92 (2005).
Häring, M. et al. Morphology and genome organization of the virus PSV of the hyperthermophilic archaeal genera Pyrobaculum and Thermoproteus: a novel virus family, the Globuloviridae. Virology 323, 233–242 (2004).
Wiedenheft, B. et al. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J. Virol. 78, 1954–1961 (2004). This paper compares four complete genomes sequenced from archaeal viruses with identical morphologies and infecting similar hosts, but isolated from four different locations around the world.
Rice, G. et al. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc. Natl Acad. Sci. USA 101, 7716–7720 (2004).
Xiang, X. et al. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus–host interactions and genomic features. J. Virol. 79, 8677–8686 (2005).
Häring, M. et al. Structure and genome organization of AFV2, a novel archaeal lipothrixvirus with unusual terminal and core structures. J. Bacteriol. 187, 3855–3858 (2005).
Arnold, H. P. et al. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267, 252–266 (2000).
Häring, M. et al. Independent virus development outside a host. Nature 436, 1101–1102 (2005). This paper, and the supplementary data, describe a virus that undergoes extracellular maturation and causes lysis of the host cell, two characteristics not seen in other crenarchaeal viruses.
Prangishvili, D. & Garrett, R. A. Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses. Biochem. Soc. Trans. 32, 204–208 (2004).
Prangishvili, D., Stedman, K. & Zillig, W. Viruses of the extremely thermophilic archaeon Sulfolobus. Trends Microbiol. 9, 39–43 (2001).
Prangishvili, D. Evolutionary insights from studies on viruses of hyperthermophilic archaea. Res. Microbiol. 154, 289–294 (2003).
Snyder, J. C. et al. Viruses of hyperthermophilic Archaea. Res. Microbiol. 154, 474–482 (2003).
Dyall-Smith, M., Tang, S. & Bath, C. Haloarchaeal viruses: how diverse are they? Res. Microbiol. 154, 309–313 (2003). This is a review of the diversity of viruses infecting the haloarchaea which describes the viruses infecting the other major group of Archaea, the Euryarchaeota.
Takai, K. & Sako, Y. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol. Ecol. 28, 177–188 (1999).
Takai, K. & Horikoshi, K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66, 5066–5072 (2000).
Spear, J. R., Walker, J. J., McCollom, T. M. & Pace, N. R. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl Acad. Sci. USA 102, 2555–2560 (2005).
Brock, T. D., Brock, K. M., Belly, R. T. & Weiss, R. L. Sulfolobus: new genus of sulfur-oxidizing bacteria living at low pH and high-temperature. Archiv. Fur Mikrobiologie 84, 54–68 (1972).
Segerer, A., Neuner, A., Kristjansson, J. K. & Stetter, K. O. Acidianus infernus gen. nov., sp. nov., and Acidianus brierleyi Comb. nov., facultatively aerobic, extremely acidophilic thermophilic sulfur-metabolizing Archaebacteria. Int. J. System. Bacteriol. 36, 559–564 (1986).
Itoh, T., Suzuki, K. & Nakase, T. Vulcanisaeta distributa gen. nov., sp nov., and Vulcanisaeta souniana sp. nov., novel hyperthermophilic rod-shaped crenarchaeotes isolated from hot springs in Japan. Int. J. System. Evol. Microbiol. 52, 1097–1104 (2002).
Svetlichnyi, V. A., Slesarev, A. I., Svetlichnaya, T. P. & Zavarzin, G. A. Caldococcus litoralis gen. nov., sp. nov., a new marine, extremely thermophilic, sulfur-reducing Archaebacterium. Microbiology 56, 658–664 (1987).
Segerer, A. H., Trincone, A., Gahrtz, M. & Stetter, K. O. Stygiolobus azoricus gen. nov., sp. nov., represents a novel genus of anaerobic, extremely thermoacidophilic archaebacteria of the order Sulfolobales. Int. J. System. Bacteriol. 41, 495–501 (1991).
Huber, G., Spinnler, C., Gambacorta, A. & Stetter, K. O. Metallosphaera sedula gen. nov. and sp. nov., represents a new genus of aerobic, metal-mobilizing, thermoacidophilic Archaebacteria. System. Appl. Microbiol. 12, 38–47 (1989).
Zillig, W. et al. Desulfurococcaceae, the 2nd family of the extremely thermophilic, anaerobic, sulfur-respiring Thermoproteales. Zentralbl. Bakteriol. Mikrobiol. Hyg. [A] 3, 304–317 (1982).
Hensel, R. et al. Sulfophobococcus zilligii gen. nov., sp. nov., a novel hyperthermophilic archaeum isolated from hot alkaline springs of Iceland. System. Appl. Microbiol. 20, 102–110 (1997).
Itoh, T., Suzuki, K. & Nakase, T. Thermocladium modestius gen. nov., sp. nov., a new genus of rod-shaped, extremely thermophilic crenarchaeote. Int. J. System. Bacteriol. 48, 879–887 (1998).
Itoh, T., Suzuki, K., Sanchez, P. C. & Nakase, T. Caldivirga maquilingensis gen. nov., sp nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int. J. System. Bacteriol. 49, 1157–1163 (1999).
Huber, R., Kristjansson, J. K. & Stetter, K. O. Pyrobaculum gen. nov., a new genus of neutrophilic, rod-shaped Archaebacteria from continental solfataras growing optimally at 100°C. Archiv. Microbiol. 149, 95–101 (1987).
Zillig, W. et al. The Archaebacterium Thermofilum pendens represents, a novel genus of the thermophilic, anaerobic sulfur respiring Thermoproteales. System. Appl. Microbiol. 4, 79–87 (1983).
Zillig, W., Tu, J. & Holz, I. Thermoproteales — a 3rd order of thermoacidophilic Archaebacteria. Nature 293, 85–86 (1981).
Itoh, T., Suzuki, K., Sanchez, P. C. & Nakase, T. Caldisphaera lagunensis gen. nov., sp. nov., a novel thermoacidophilic crenarchaeote isolated from a hot spring at Mt. Maquiling, Philippines. Int. J. System. Evol. Microbiol. 53, 1149–1154 (2003).
Snyder, J. C. Virus dynamics, archaeal populations, and water chemistry of three acidic hot springs in Yellowstone National Park. Ph.D. Thesis, Univ. Montana State. (2005).
Breitbart, M., Wegley, L., Leeds, S., Schoenfeld, T. & Rohwer, F. Phage community dynamics in hot springs. Appl. Environ. Microbiol. 70, 1633–1640 (2004).
Karner, M. B., Delong, E. F. & Karl, D. M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510 (2001).
Simon, H. M. et al. Cultivation of mesophilic soil crenarchaeotes in enrichment cultures from plant roots. Appl. Environ. Microbiol. 71, 4751–4760 (2005).
Pinar, G., Gurtner, C., Lubitz, W. & Rolleke, S. Identification of archaea in objects of art by denaturing gradient gel electrophoresis analysis and shotgun cloning. Methods Enzymol. 336, 356–366 (2001).
Rieu-Lesme, F., Delbes, C. & Sollelis, L. Recovery of partial 16S rDNA sequences suggests the presence of Crenarchaeota in the human digestive ecosystem. Curr. Microbiol. 51, 317–321 (2005).
Buckley, D. H., Graber, J. R. & Schmidt, T. M. Phylogenetic analysis of non-thermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl. Environ. Microbiol. 64, 4333–4339 (1998).
Ochsenreiter, T., Selezi, D., Quaiser, A., Bonch-Osmolovskaya, L. & Schleper, C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5, 787–797 (2003).
Massana, R., Murray, A. E., Preston, C. M. & Delong, E. F. Vertical distribution and phylogenetic characterization of marine planktonic archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63, 50–56 (1997).
Murray, A. E. et al. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl. Environ. Microbiol. 64, 2585–2595 (1998).
Geslin, C. et al. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, Pyrococcus abyssi. J. Bacteriol. 185, 3888–3894 (2003).
Maniloff, J. & Ackermann, H. W. Taxonomy of bacterial viruses: Establishment of tailed virus genera and the order Caudovirales. Archiv. Virol. 143, 2051–2063 (1998).
Rice, G. et al. Viruses from extreme thermal environments. Proc. Natl Acad. Sci. USA 98, 13341–13345 (2001). This paper describes the diversity of viruses detected from enrichment cultures from Yellowstone hot spring samples.
Rachel, R. et al. Remarkable morphological diversity of viruses and virus-like particles in hot terrestrial environments. Archiv. Virol. 147, 2419–2429 (2002).
Snyder, J. C. et al. Effects of culturing on the population structure of a hyperthermophilic virus. Microb. Ecol. 48, 561–566 (2004).
Janekovic, D. et al. TTV1, TTV2 and TTV3, a family of viruses of the extremely thermophilic, anaerobic, sulfur reducing archaebacterium Thermoproteus tenax. Mol. Gen. Genet. 192, 39–45 (1983).
Prangishvili, D. et al. A novel virus family, the Rudiviridae: structure, virus–host interactions and genome variability of the Sulfolobus viruses SIRV1 and SIRV2. Genetics 152, 1387–1396 (1999).
Martin, A. et al. SAV-1, a temperate UV-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius Isolate B-12. EMBO J. 3, 2165–2168 (1984).
Stedman, K. M. et al. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res. Microbiol. 154, 295–302 (2003).
Bath, C. & Dyall-Smith, M. L. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J. Virol. 72, 9392–9395 (1998).
Wood, A. G., Whitman, W. B. & Konisky, J. Isolation and characterization of an archaebacterial virus-like particle from Methanococcus voltae A3. J. Bacteriol. 171, 93–98 (1989).
Khayat, R. et al. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc. Natl Acad. Sci. USA 102, 18944–18949 (2005). A recent paper describing the crystal structure of the major capsid protein from STIV which adds further support to the hypothesis that icosahedral dsDNA viruses share a common ancestry.
Maaty, W. S. A. et al. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among dsDNA viruses from all domains of life. J. Virol. (In the press).
Porter, K. et al. SH1: a novel, spherical halovirus isolated from an Australian hypersaline lake. Virology 335, 22–33 (2005).
Bamford, D. H. et al. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J. Virol. 79, 9097–9107 (2005).
Baker, M. L., Jiang, W., Rixon, F. J. & Chiu, W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 79, 14967–14970 (2005).
Duda, R. L., Hendrix, R. W., Huang, W. M. & Conway, J. F. Shared architecture of bacteriophage SPO1 and herpesvirus capsids. Curr. Biol. 16, R11–R13 (2006).
Mallick, P., Boutz, D. R., Eisenberg, D. & Yeates, T. O. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc. Natl Acad. Sci. USA 99, 9679–9684 (2002).
Nadal, M., Mirambeau, G., Forterre, P., Reiter, W. & Duguet, M. Positively supercoiled DNA in a virus-like particle of an archaebacterium. Nature 321, 256–258 (1986).
Forterre, P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 18, 236–237 (2002).
Peng, X. et al. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291, 226–234 (2001).
Grogan, D. W. Hyperthermophiles and the problem of DNA instability. Mol. Microbiol. 28, 1043–1049 (1998).
Kraft, P. et al. Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein. J. Virol. 78, 11544–11550 (2004).
Kraft, P. et al. Structure of D-63 from Sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function. J. Virol. 78, 7438–7442 (2004).
Hendrix, R. W., Smith, M. C. M., Burns, R. N., Ford, M. E. & Hatfull, G. F. Evolutionary relationships among diverse bacteriophages: all the world's a phage. Proc. Natl Acad. Sci. USA 96, 2192–2197 (1999).
del Solar, G., Giraldo, R., Ruiz-Echevarria, M. J., Espinosa, M. & Diaz-Orejas, R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62, 434–464 (1998).
She, Q., Shen, B. & Chen, L. Archaeal integrases and mechanisms of gene capture. Biochem. Soc. Trans. 32, 222–226 (2004).
Yeats, S., Mcwilliam, P. & Zillig, W. A Plasmid in the archaebacterium Sulfolobus acidocaldarius. EMBO J. 1, 1035–1038 (1982).
Swalla, B. M., Gumport, R. I. & Gardner, J. F. Conservation of structure and function among tyrosine recombinases: homology-based modeling of the λ integrase core-binding domain. Nucleic Acids Res. 31, 805–818 (2003).
Reiter, W. D., Palm, P., Yeats, S. & Zillig, W. Gene expression in Archaebacteria: physical mapping of constitutive and UV-inducible transcripts from the Sulfolobus virus-like particle SSV1. Mol. Gen. Genet. 209, 270–275 (1987).
Kessler, A., Brinkman, A. B., van der Oost, J. & Prangishvili, D. Transcription of the rod-shaped viruses SIRV1 and SIRV2 of the hyperthermophilic archaeon Sulfolobus. J. Bacteriol. 186, 7745–7753 (2004).
Schleper, C., Kubo, K. & Zillig, W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl Acad. Sci. USA 89, 7645–7649 (1992).
Liu, D. X. & Huang, L. Induction of the Sulfolobus shibatae virus SSV1 DNA replication by mitomycin C. Chinese Sci. Bull. 47, 923–927 (2002).
Zillig, W. et al. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. System. Appl. Microbiol. 16, 609–628 (1994).
Palm, P. et al. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185, 242–250 (1991).
Blum, H., Zillig, W., Mallok, S., Domdey, H. & Prangishvili, D. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology 281, 6–9 (2001).
Ackermann, H. W. Bacteriophage observations and evolution. Res. Microbiol. 154, 245–251 (2003).
Whitaker, R. J., Grogan, D. W. & Taylor, J. W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301, 976–978 (2003).
Suttle, C. A. Viruses in the sea. Nature 437, 356–361 (2005).
Acknowledgements
We would like to acknowledge the pioneering work of W. Zillig in the study of archaeal viruses. Much of our work described in this review was supported by funding from the National Science Foundation and NASA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome
Entrez Genome Project
FURTHER INFORMATION
Universal Virus Database of the International Committee on Taxonomy of Viruses
Glossary
- Methanogen
-
Anaerobic archaeon that produces methane as a waste product of autotrophic metabolism.
- Halophile
-
An aerobic organism that requires salt for survival. Extreme halophiles (environmental salt concentration >15%) are all archaea.
- Thermophile
-
An organism with an optimal growth rate above 50°C. Organisms with optimal growth rates at temperatures greater than 75°C are classified as hyperthermophiles.
- Polyamine
-
An organic compound synthesized in cells and required for growth. These compounds have two or more amine groups and are positively charged, enabling them to bind DNA.
- Glycosyltransferase
-
An enzyme that tranfers glycosyl (carbohydrate radical) from one compound to another. There are several families of these enzymes.
- Tyrosine recombinase
-
A diverse group of proteins involved in recombination between DNA at specific sites. Functions include integration of virus genomes, relaxation of DNA supercoils, conjugation and genome separation during cell division.
- Holliday junction resolvase
-
An enzyme that cleaves concatamer junctions to create linear duplex DNA during genome replication in poxviruses.
Rights and permissions
About this article
Cite this article
Ortmann, A., Wiedenheft, B., Douglas, T. et al. Hot crenarchaeal viruses reveal deep evolutionary connections. Nat Rev Microbiol 4, 520–528 (2006). https://doi.org/10.1038/nrmicro1444
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1444
This article is cited by
-
Diversity, evolutionary contribution and ecological roles of aquatic viruses
Science China Life Sciences (2018)
-
Repression of RNA polymerase by the archaeo-viral regulator ORF145/RIP
Nature Communications (2016)
-
When Competing Viruses Unify: Evolution, Conservation, and Plasticity of Genetic Identities
Journal of Molecular Evolution (2015)
-
Molecular biology of fuselloviruses and their satellites
Extremophiles (2014)
-
Archaea — timeline of the third domain
Nature Reviews Microbiology (2011)