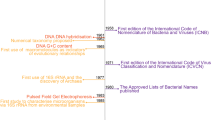
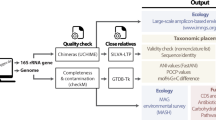
Abstract
There is no widely accepted concept of species for prokaryotes, and assignment of isolates to species is based on measures of phenotypic or genome similarity. The current methods for defining prokaryotic species are inadequate and incapable of keeping pace with the levels of diversity that are being uncovered in nature. Prokaryotic taxonomy is being influenced by advances in microbial population genetics, ecology and genomics, and by the ease with which sequence data can be obtained. Here, we review the classical approaches to prokaryotic species definition and discuss the current and future impact of multilocus nucleotide-sequence-based approaches to prokaryotic systematics. We also consider the potential, and difficulties, of assigning species status to biologically or ecologically meaningful sequence clusters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Stackebrandt, E. et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52, 1043–1047 (2002).
Vandamme, P. et al. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60, 407–438 (1996).
Amann, R. I., Ludwig, W. & Schleifer, K. H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169 (1995).
Cohan, F. M. What are bacterial species? Annu. Rev. Microbiol. 56, 457–487 (2002).
Woese, C. R. The Use of Ribosomal RNA in Reconstructing Evolutionary Relationships Among Bacteria. in Evolution at the Molecular Level (eds Selander, R. K., Clark, A. G. & Whittam, T. S.) Ch. 1, 1–24 (Sinauer Associates Inc., Sunderland, 1991).
Fox, G. E., Wisotzkey, J. D. & Jurtshuk, P. Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42, 166–170 (1992).
Stackebrandt, E. & Goebel, B. M. A place for DNA–DNA reassociation and 16S ribosomal-RNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849 (1994).
Gogarten, J. P., Doolittle, W. F. & Lawrence, J. G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19, 2226–2238 (2002).
Boucher, Y., Douady, C. J., Sharma, A. K., Kamekura, M. & Doolittle, W. F. Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J. Bacteriol. 186, 3980–3990 (2004).
Maiden, M. C. J. et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl Acad. Sci. USA 95, 3140–3145 (1998).
Cooper, J. E. & Feil, E. J. Multilocus sequence typing — what is resolved? Trends Microbiol. 12, 373–377 (2004).
Zeigler, D. R. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int. J. Syst. Evol. Microbiol. 53, 1893–1900 (2003).
Santos, S. R. & Ochman, H. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ. Microbiol. 6, 754–759 (2004).
Thompson, F. L. et al. Phylogeny and molecular identification of vibrios based on multilocus sequence analysis. Appl. Environ. Microbiol. (in the press).
Devulder, G., Perouse de Montclos, M. & Flandrois, J. P. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55, 293–302 (2005).
Kotetishvili, M. et al. Multilocus sequence typing for studying genetic relationships among Yersinia species. J. Clin. Microbiol. 43, 2674–2684 (2005).
Priest, F. G., Barker, M., Baillie, L. W. J., Holmes, E. C. & Maiden, M. C. J. Population structure and evolution of the Bacillus cereus group. J. Bacteriol. 186, 7959–7970 (2004).
Godoy, D. et al. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41, 2068–2079 (2003).
Feldgarden, M., Byrd, N. & Cohan, F. M. Gradual evolution in bacteria: evidence from Bacillus systematics. Microbiology 149, 3565–3573 (2003).
Welch, R. A. et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA 99, 17020–17024 (2002).
Schloter, M., Lebuhn, M., Heulin, T. & Hartmann, A. Ecology and evolution of bacterial microdiversity. FEMS Microbiol. Rev. 24, 647–660 (2000).
Jolley, K. A. et al. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 38, 4492–4498 (2000).
Enright, M. C., Spratt, B. G., Kalia, A., Cross, J. H. & Bessen, D. E. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69, 2416–2427 (2001).
Sreevatsan, S. et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl Acad. Sci. USA 94, 9869–9874 (1997).
Godreuil, S., Cohan, F. M., Shah, H. & Tibayrenc, M. Which species concept for pathogenic bacteria?: an e-debate. Infect. Genet. Evol. 20 Jan 2005 (doi/10.1016/j.meegid.2004.03.004).
de Queiroz, K. in Endless Forms: Species and Speciation (eds Howard, D. J. & Berlocher, S. H.) Ch. 5, 57–75 (Oxford University Press, Oxford, 1998).
Ward, D. M. & Cohan, F. M. Microbial Diversity in Hot Spring Cyanobacterial Mats: Pattern and Prediction. in Geothermal Biology and Geochemistry in Yellowstone National Park (eds Inskeep, W. P. & McDermott, T.) in the press (Thermal Biology Institute, Bozeman).
Hanage, W. P., Fraser, C. & Spratt, B. G. Fuzzy species among recombinogenic bacteria. BMC Biol. 3, 6 (2005).
Feil, E. J. et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl Acad. Sci. USA 98, 182–187 (2001).
Field, D., Feil, E. J. & Wilson, G. Databases and software for the comparative genomic study of collections of genomes. Microbiology (in the press).
Welch, D. F. Applications of cellular fatty acid analysis. Clin. Microbiol. Rev. 4, 422–438 (1991).
Vos, P. et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414 (1995).
van Belkum, A. DNA fingerprinting of medically important microorganisms by use of PCR. Clin. Microbiol. Rev. 7, 174–184 (1994).
Coenye, T. & Vandamme, P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5, 719–729 (2003).
Holden, M. T. G. et al. Genornic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl Acad. Sci. USA 101, 14240–14245 (2004).
Nierman, W. C. et al. Structural flexibility in the Burkholderia mallei genome. Proc. Natl Acad. Sci. USA 101, 14246–14251 (2004).
Brett, P. J., Deshazer, D., & Woods, D. E. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48, 317–320 (1998).
Rogul, M., Brendle, J. J., Haapala, D. K. & Alexander, A. D. Nucleic acid similarities among Pseudomonas pseudomallei, Pseudomonas multivorans, and Actinobacillus mallei. J. Bacteriol. 101, 827–835 (1970).
Yabuuchi, E. et al. Burkholderia uboniae sp. nov., L-arabinose-assimilating but different from Burkholderia thailandensis and Burkholderia vietnamiensis. Microbiol. Immunol. 44, 307–317 (2000).
Cohan, F. M. Concepts of Bacterial Biodiversity for the Age of Genomics. in Microbial Genomes (eds Fraser, C. M., Read, T. D. & Nelson, K. E.) Ch. 11, 175–194 (Humana Press, Totowa, 2004).
Lawrence, J. G. Gene transfer in bacteria: speciation without species. Theor. Popul. Biol. 61, 449–460 (2002).
Palys, T., Nakamura, L. K. & Cohan, F. M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47, 1145–1156 (1997).
Papke, R. T., Ramsing, N. B., Bateson, M. M. & Ward, D. M. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5, 650–659 (2003).
Fraser, C., Hanage, W. P. & Spratt, B. G. Neutral microepidemic evolution of bacterial pathogens? Proc. Natl Acad. Sci. USA 102, 1968–1973 (2005).
Cohan, F. M. Does recombination constrain neutral divergence among bacterial taxa. Evolution 49, 164–175 (1995).
Keim, P. et al. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4, 205–213 (2004).
Dunlap, P. V. and Ast, J. C. Genomic and phylogenetic characterization of luminous bacteria symbiotic with the deep-sea fish Chlorophthalmus albatrossis (Aulopiformes: Chlorophthalmidae). Appl. Environ. Microbiol. 71, 930–939 (2005).
Acknowledgements
This paper is the outcome of a workshop entitled ‘The prokaryotic species: genome plasticity, microevolution and taxonomy’, organized by F. Thompson and J. Swings in Ghent, Belgium, 25 October 2004, and has the intention to stir up the interdisciplinary debate on the prokaryotic species. We thank all participants as well as D. Mazel, G. Manfio and T. Iida for their contributions. T.C., P.V. and J.S. are indebted to the Fund for Scientific Research — Flanders (Belgium) for a position as postdoctoral fellow and research funding, respectively. F.M.C. acknowledges funding from the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- MLST
-
Multilocus sequence typing, a method for the genotypic characterization of prokaryotes at the infraspecific level, using the allelic mismatches of a small number (usually 7) of housekeeping genes. Designed as a tool in molecular epidemiology and used for recognizing distinct strains within named species.
- MLSA
-
Multilocus sequence analysis, a method for the genotypic characterization of a more diverse group of prokaryotes (including entire genera) using the sequences of multiple protein-coding genes.
- SPECIES CONCEPT
-
A framework to understand how and why an observer can sort organisms into species; that is, what kind of unit do we think the term species embraces, and what characteristics are shared between all members of a species.
- SPECIES DEFINITION
-
A more practical outline of how to assign isolates to a named species or identify new species.
Rights and permissions
About this article
Cite this article
Gevers, D., Cohan, F., Lawrence, J. et al. Re-evaluating prokaryotic species. Nat Rev Microbiol 3, 733–739 (2005). https://doi.org/10.1038/nrmicro1236
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1236
This article is cited by
-
Enhancing mass production of Heterorhabditis bacteriophora: influence of different bacterial symbionts (Photorhabdus spp.) and inoculum age on dauer juvenile recovery
World Journal of Microbiology and Biotechnology (2024)
-
Distributed genotyping and clustering of Neisseria strains reveal continual emergence of epidemic meningococcus over a century
Nature Communications (2023)
-
Bacterial motility can govern the dynamics of antibiotic resistance evolution
Nature Communications (2023)
-
Genomic insights into zoonotic transmission and antimicrobial resistance in Campylobacter jejuni from farm to fork: a one health perspective
Gut Pathogens (2022)
-
Intraspecies characterization of bacteria via evolutionary modeling of protein domains
Scientific Reports (2022)