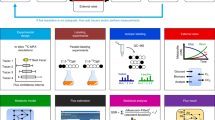
Key Points
-
The metabolome can be defined as the quantitative complement of low-molecular-weight metabolites present in a cell under a given set of physiological conditions. As changes in a cell's physiology as a result of gene deletion or overexpression are amplified through the hierarchy of the transcriptome and the proteome, it can be argued that such changes are more easily measurable through the metabolome, and that the metabolome is more sensitive to perturbations than is either the transcriptome or proteome.
-
High-throughput measurement of a complete set of intracellular microbial metabolites would require the development of an accurate, simple and rapid method with a broad dynamic range suitable for detecting and quantifying large numbers of metabolites that can be present at different concentrations.
-
Instead, the global exometabolome – or secreted metabolites - can be analysed using metabolic footprinting, a convenient and reproducible technique to monitor metabolites consumed from, and secreted into, the growth medium by batch cultures of yeast using direct-injection mass spectrometry. The principles of the technique, and results obtained using it, are described.
-
Metabolic footprinting can therefore be used to generate metabolomic data, which, in the era of systems biology, should be able to be integrated with transcriptomic and proteomic data. The current available data models and data standards for systems biology, and the need for the metabolomics community to follow the lead of transcriptomics and proteomics researchers, are also discussed.
Abstract
One element of classical systems analysis treats a system as a black or grey box, the inner structure and behaviour of which can be analysed and modelled by varying an internal or external condition, probing it from outside and studying the effect of the variation on the external observables. The result is an understanding of the inner make-up and workings of the system. The equivalent of this in biology is to observe what a cell or system excretes under controlled conditions — the 'metabolic footprint' or exometabolome — as this is readily and accurately measurable. Here, we review the principles, experimental approaches and scientific outcomes that have been obtained with this useful and convenient strategy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Oliver, S. G., Winson, M. K., Kell, D. B. & Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 16, 373–378 (1998). First use of the term 'metabolome' in the literature.
Fiehn, O., Kloska, S. & Altmann, T. Integrated studies on plant biology using multiparallel techniques. Curr. Opin. Biotechnol. 12, 82–86 (2001).
Fiehn, O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genomics 2, 155–168 (2001).
Harrigan, G. G. & Goodacre, R. (eds) Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis (Kluwer Academic Publishers, Boston, 2003). The first book on metabolomics.
Weckwerth, W. Metabolomics in systems biology. Annu. Rev. Plant Biol. 54, 669–689 (2003).
Goodacre, R., Vaidyanathan, S., Dunn, W. B., Harrigan, G. G. & Kell, D. B. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 22, 245–252 (2004).
Nicholson, J. K., Holmes, E., Lindon, J. C. & Wilson, I. D. The challenges of modeling mammalian biocomplexity. Nature Biotechnol. 22, 1268–1274 (2004). Stresses the role of intestinal microorganisms in contributing to the human metabolome.
van der Greef, J., Stroobant, P. & van der Heijden, R. The role of analytical sciences in medical systems biology. Curr. Opin. Chem. Biol. 8, 559–565 (2004). The importance of analytical sciences to omics and systems biology.
Kell, D. B. Metabolomics and systems biology: making sense of the soup. Curr. Opin. Microbiol. 7, 296–307 (2004). Metabolomic data as an input to systems biology models.
Mendes, P., Kell, D. B. & Westerhoff, H. V. Why and when channeling can decrease pool size at constant net flux in a simple dynamic channel. Biochim. Biophys. Acta 1289, 175–186 (1996).
Oliver, S. G. Yeast as a navigational aid in genome analysis. Microbiology 143, 1483–1487 (1997).
Raamsdonk, L. M. et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nature Biotechnol. 19, 45–50 (2001).
ter Kuile, B. H. & Westerhoff, H. V. Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett. 500, 169–171 (2001).
Fell, D. A. Understanding the Control of Metabolism (Portland Press, London, 1996). Excellent introduction to metabolic control analysis.
Heinrich, R. & Schuster, S. The Regulation of Cellular Systems (Chapman & Hall, New York, 1996).
Kell, D. B. & Mendes, P. in Technological and Medical Implications of Metabolic Control Analysis (eds Cornish–Bowden, A. & Cárdenas, M. L.) 3–25 (Kluwer Academic Publishers, Dordrecht, 2000).
Urbanczyk-Wochniak, E. et al. Parallel analysis of transcript and metabolic profiles: a new approach in systems biology. EMBO Rep. 4, 989–993 (2003).
Kell, D. B., Kaprelyants, A. S. & Grafen, A. On pheromones, social behaviour and the functions of secondary metabolism in bacteria. Trends Ecol. Evol. 10, 126–129 (1995).
De Koning, W. & van Dam, K. A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal. Biochem. 204, 118–123 (1992).
Teusink, B., Baganz, F., Westerhoff, H. V. & Oliver, S. G. in Methods in Microbiology: Yeast Gene Analysis (eds Tuite, M. F. & Brown, A. J. P.) 297–336 (Academic Press, London, 1998).
Gonzalez, B., Francois, J. & Renaud, M. A rapid and reliable method for metabolite extraction in yeast using boiling buffered ethanol. Yeast 13, 1347–1355 (1997).
Castrillo, J. I., Hayes, A., Mohammed, S., Gaskell, S. J. & Oliver, S. G. An optimized protocol for metabolome analysis in yeast using direct infusion electrospray mass spectrometry. Phytochem. 62, 929–937 (2003).
Lindon, J. C., Nicholson, J. K. & Everett, J. R. NMR spectroscopy of biofluids. Annu. Rep. NMR Spectrosc. 38, 1–88 (1999).
Soga, T. et al. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2, 488–494 (2003).
Greenaway, W., May, J., Scaysbrook, T. & Whatley, F. R. Identification by gas chromatography-mass spectrometry of 150 compounds in propolis. Z. Naturforsch. 46, 111–121 (1991). A pioneering paper in plant metabolomics.
Fiehn, O. et al. Metabolite profiling for plant functional genomics. Nature Biotechnol. 18, 1157–1161 (2000). An important paper that initiated the modern era of plant metabolomics.
Roessner, U. et al. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13, 11–29 (2001).
O'Hagan, S., Dunn, W. B., Brown, M., Knowles, J. D. & Kell, D. B. Closed-loop, multiobjective optimisation of analytical instrumentation: gas-chromatography-time-of-flight mass spectrometry of the metabolomes of human serum and of yeast fermentations. Anal. Chem. 77, 290–303 (2005). Explicit recognition of the need for optimization in metabolomics, and a new way of doing it.
van Eijk, H. M. H., Rooyakkers, D. R., Soeters, P. B. & Deutz, N. E. P. Determination of amino acid isotope enrichment using liquid chromatography-mass spectrometry. Anal. Biochem. 271, 8–17 (1999).
Buchholz, A., Takors, R. & Wandrey, C. Quantification of intracellular metabolites in Escherichia coli K12 using liquid chromatographic–electrospray ionization tandem mass spectrometric techniques. Anal. Biochem. 295, 129–137 (2001).
Lenz, E. M., Bright, J., Knight, R., Wilson, I. D. & Major, H. A metabonomic investigation of the biochemical effects of mercuric chloride in the rat using 1H NMR and HPLC-TOF/MS: time dependent changes in the urinary profile of endogenous metabolites as a result of nephrotoxicity. Analyst 129, 535–541 (2004).
Colón, L. A., Cintron, J. M., Anspach, J. A., Fermier, A. M. & Swinney, K. A. Very high pressure HPLC with 1 mm id columns. Analyst 129, 503–504 (2004).
Welch, G. R. & Easterby, J. S. Metabolic channeling versus free diffusion: transition-time analysis. Trends Biochem. Sci. 19, 193–197 (1994).
Rashed, M. S. et al. Screening blood spots for inborn errors of metabolism by electrospray tandem mass spectrometry with a microplate batch process and a computer algorithm for automated flagging of abnormal profiles. Clin. Chem. 43, 1129–1141 (1997).
Nicholson, J. K. & Wilson, I. D. Understanding 'global' systems biology: metabonomics and the continuum of metabolism. Nature Rev. Drug Disc. 2, 668–676 (2003).
Barker, R. L., Gracey, D. E. F., Irwin, A. J., Pipasts, P. & Leiska, E. Liberation of staling aldehydes during storage of beer. J. Inst. Brew. 89, 411–415 (1983).
Lynch, P. A. & Seo, C. W. Ethylene production in staling beer. J. Food Sci. 52, 1270–1272 (1987).
Rinas, U., Hellmuth, K., Kang, R. J., Seeger, A. & Schlieker, H. Entry of Escherichia coli into stationary phase is indicated by endogenous and exogenous accumulation of nucleobases. Appl. Env. Microbiol. 61, 4147–4151 (1995).
Shiga, Y., Mizuno, H. & Akanuma, H. Conditional synthesis and utilization of 1,5-anhydroglucitol in Escherichia coli. J. Bacteriol. 175, 7138–7141 (1993).
Hayashida, Y., Kuriyama, H., Nishimura, K. & Slaughter, J. C. Production of 4-hydroxyfuranones in simple media by fermentation. Biotechnol. Lett. 21, 505–509 (1999).
Slaughter, J. C. The naturally occurring furanones: formation and function from pheromone to food. Biol. Rev. Camb. Philos. Soc. 74, 259–276 (1999).
Kaprelyants, A. S. & Kell, D. B. Do bacteria need to communicate with each other for growth? Trends Microbiol. 4, 237–242 (1996).
Fuqua, C., Winans, S. C. & Greenberg, E. P. Census and consensus in bacterial ecosystems: the LuxR–LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50, 727–751 (1996).
Mukamolova, G. V., Kaprelyants, A. S., Young, D. I., Young, M. & Kell, D. B. A bacterial cytokine. Proc. Natl Acad. Sci. USA 95, 8916–8921 (1998).
Weichart, D. H. & Kell, D. B. Characterization of an autostimulatory substance produced by Escherichia coli. Microbiology 147, 1875–1885 (2001).
Firstenberg-Eden, R. & Eden, G. Impedance Microbiology (Research Studies Press, Letchworth, 1984).
Holms, H. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol. Rev. 19, 85–116 (1996).
Wittmann, C. & Heinzle, E. MALDI-TOF MS for quantification of substrates and products in cultivations of Corynebacterium glutamicum. Biotechnol. Bioeng. 72, 642–647 (2001).
Devlin, J. P. (ed.) High Throughput Screening: The Discovery of Bioactive Substances (Marcel Dekker, New York, 1997).
Abel, C. B. L. et al. Characterization of metabolites in intact Streptomyces citricolor culture supernatants using high-resolution nuclear magnetic resonance and directly coupled high-pressure liquid chromatography-nuclear magnetic resonance spectroscopy. Anal. Biochem. 270, 220–230 (1999).
Bubb, W. A. et al. Heteronuclear NMR studies of metabolites produced by Cryptococcus neoformans in culture media: identification of possible virulence factors. Magn. Res. Med. 42, 442–453 (1999).
Bachinger, T. & Mandenius, C. F. Searching for process information in the aroma of cell cultures. Trends Biotechnol. 18, 494–500 (2000).
Gibson, T. D. et al. Detection and simultaneous identification of microorganisms from headspace samples using an electronic nose. Sens. Actuators B 44, 413–422 (1997).
Allen, J. K. et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nature Biotechnol. 21, 692–696 (2003). The original paper describing metabolic footprinting.
Fiehn, O. Metabolomics: the link between genotypes and phenotypes. Plant Mol. Biol. 48, 155–171 (2002).
Neijssel, O. M. & Tempest, D. W. The role of energy-splitting reactions in the growth of Klebsiella aerogenes NCTC 418 in aerobic chemostat culture. Arch. Microbiol. 110, 305–311 (1976).
Lazebnik, Y. Can a biologist fix a radio? — Or, what I learned while studying apoptosis. Cancer Cell 2, 179–182 (2002).
Westerhoff, H. V. & Kell, D. B. Matrix method for determining the steps most rate-limiting to metabolic fluxes in biotechnological processes. Biotechnol. Bioeng. 30, 101–107 (1987).
Kaderbhai, N. N., Broadhurst, D. I., Ellis, D. I., Goodacre, R. & Kell, D. B. Functional genomics via metabolic footprinting: monitoring metabolite secretion by Escherichia coli tryptophan metabolism mutants using FT-IR and direct injection electrospray mass spectrometry. Comp. Funct. Genomics 4, 376–391 (2003).
Allen, J. et al. Discrimination of modes of action of antifungal substances by use of metabolic footprinting. Appl. Environ. Microbiol. 70, 6157–6165 (2004).
Langdon, W. B. Genetic Programming and Data Structures: Genetic Programming + Data Structures = Automatic Programming! (Kluwer Academic Publishers, Boston, 1998).
Kell, D. B. Genotype–phenotype mapping: genes as computer programs. Trends Genet. 18, 555–559 (2002).
Koza, J. R. et al. Genetic Programming: Routine Human–Competitive Machine Intelligence (Kluwer Academic Publishers, New York, 2003).
Schreiber, S. L. Chemical genetics resulting from a passion for synthetic organic chemistry. Bioorg. Med. Chem. 6, 1127–1152 (1998).
Huang, J. et al. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl Acad. Sci. USA 101, 16594–16599 (2004).
Brown, M. et al. A metabolome pipeline: from concept to data to knowledge. Metabolomics 1, 35–46 (2005).
Camacho, D., de la Fuente, A. & Mendes, P. The origins of correlations in metabolomics data. Metabolomics 1, 53–63 (2005).
Kose, F., Weckwerth, W., Linke, T. & Fiehn, O. Visualizing plant metabolomic correlation networks using clique-metabolite matrices. Bioinformatics 17, 1198–1208 (2001).
Fiehn, O. Metabolic networks of Cucurbita maxima phloem. Phytochemistry 62, 875–886 (2003).
Steuer, R., Kurths, J., Fiehn, O. & Weckwerth, W. Observing and interpreting correlations in metabolomic networks. Bioinformatics 19, 1019–1026 (2003).
Ravasz, E., Somera, A. L., Mongru, D. A., Oltvai, Z. N. & Barabási, A. L. Hierarchical organization of modularity in metabolic networks. Science 297, 1551–1555 (2002).
Csete, M. E. & Doyle, J. C. Reverse engineering of biological complexity. Science 295, 1664–1669 (2002). Highlights the fact that most systems-biology problems are inverse problems, in which we must move from measured variables to the inference of parameters.
Barabási, A.-L. & Oltvai, Z. N. Network biology: understanding the cell's functional organization. Nature Rev. Genet. 5, 101–113 (2004). Useful review of network biology.
Bruggeman, F. J., Westerhoff, H. V., Hoek, J. B. & Kholodenko, B. N. Modular response analysis of cellular regulatory networks. J. Theor. Biol. 218, 507–520 (2002).
Ihmels, J. et al. Revealing modular organization in the yeast transcriptional network. Nature Genet. 31, 370–377 (2002).
Han, J. D. et al. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature 430, 88–93 (2004).
Tanay, A., Sharan, R., Kupiec, M. & Shamir, R. Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genomewide data. Proc. Natl Acad. Sci. USA 101, 2981–2986 (2004).
Hughes, T. R. et al. Functional discovery via a compendium of expression profiles. Cell 102, 109–126 (2000).
Featherstone, D. E. & Broadie, K. Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network. Bioessays 24, 267–274 (2002).
Hayes, A. et al. Hybridization array technology coupled with chemostat culture: tools to interrogate gene expression in Saccharomyces cerevisiae. Methods 26, 281–290 (2002).
Vaidyanathan, S., Broadhurst, D. I., Kell, D. B. & Goodacre, R. Explanatory optimisation of protein mass spectrometry via genetic search. Anal. Chem. 75, 6679–6686 (2003).
Vaidyanathan, S., Kell, D. B. & Goodacre, R. Selective detection of proteins in mixtures using electrospray ionization mass spectrometry: influence of instrumental settings and implications for proteomics. Anal. Chem. 76, 5024–5032 (2004).
King, R. D. et al. Functional genomic hypothesis generation and experimentation by a robot scientist. Nature 427, 247–252 (2004). The 'robot scientist' strategy for principled and automated scientific hypothesis generation and testing.
Schauer, N. et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579, 1332–1337 (2005).
Marriott, P. & Shellie, R. Principles and applications of comprehensive two-dimensional gas chromatography. Trends Anal. Chem. 21, 573–583 (2002).
Wilson, I. D. & Brinkman, U. A. Hyphenation and hypernation: the practice and prospects of multiple hyphenation. J. Chromatogr. A 1000, 325–356 (2003).
Guttman, A., Varoglu, M. & Khandurina, J. Multidimensional separations in the pharmaceutical arena. Drug Discov. Today 9, 136–144 (2004).
Welthagen, W. et al. Comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry (GC × GC-TOF) for high-resolution metabolomics: biomarker discovery on spleen tissue extracts of obese NZO compared to lean C57BL/6 mice. Metabolomics 1, 65–73 (2005).
Castrillo, J. I. & Oliver, S. G. Yeast as a touchstone in post-genomic research: strategies for integrative analysis in functional genomics. J. Biochem. Mol. Biol. 37, 93–106 (2004).
Foury, F. Human genetic diseases: a cross-talk between man and yeast. Gene 195, 1–10 (1997).
Zhang, N. et al. Using yeast to place human genes in functional categories. Gene 303, 121–129 (2003).
Brazma, A. et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genet. 29, 365–371 (2001).
Spellman, P. et al. Design and implementation of microarray gene expression markup language (MAGE–ML). Genome Biol. 3, 9 (2002).
Taylor, C. F. et al. A systematic approach to modelling capturing and disseminating proteomics experimental data. Nature Biotechnol. 21, 247–254 (2003).
Garwood, K. L. et al. Pedro: a configurable data entry tool for XML. Bioinformatics 20, 2463–2465 (2004).
Garwood, K. L. et al. PEDRo: a database for storing, searching and disseminating experimental proteomics data. BMC Genomics 5 (2004).
Orchard, S., Hermjakob, H. & Apweiler, R. The proteomics standards initiative. Proteomics 3, 1374–1376 (2003).
Jenkins, H. et al. A proposed framework for the description of plant metabolomics experiments and their results. Nature Biotechnol. 22, 1601–1606 (2004). Specific schemas for evolving metabolome data standards.
Bino, R. J. et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 9, 418–425 (2004).
Kopka, J. et al. GMD@CSB. DB: the Golm metabolome database. Bioinformatics (2005).
Pedrioli, P. G. et al. A common open representation of mass spectrometry data and its application to proteomics research. Nature Biotechnol. 22, 1459–1466 (2004).
Xirasagar, S. et al. CEBS object model for systems biology data, SysBio-OM. Bioinformatics 20, 2004–2015 (2004).
Achard, F., Vaysseix, G. & Barillot, E. XML, bioinformatics and data integration. Bioinformatics 17, 115–125 (2001).
Li, X. J. et al. in Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis (eds Harrigan, G. G. & Goodacre, R.) 293–309 (Kluwer Academic Publishers, Boston, 2003).
Hucka, M. et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19, 524–531 (2003). A starting point for learning about SBML.
Finney, A. & Hucka, M. Systems biology markup language: level 2 and beyond. Biochem. Soc. Trans. 31, 1472–1473 (2003).
von Bertalanffy, L. General System Theory (George Braziller, New York, 1969).
Iberall, A. S. Toward A General Science of Viable Systems (McGraw–Hill, New York, 1972).
Ideker, T., Galitski, T. & Hood, L. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2, 343–372 (2001).
Henry, C. M. Systems biology. Chem. Eng. News 81, 45–55 (2003).
Hood, L. Systems biology: integrating technology, biology, and computation. Mech. Ageing Dev. 124, 9–16 (2003).
Kell, D. B. & Westerhoff, H. V. Metabolic control theory: its role in microbiology and biotechnology. FEMS Microbiol. Rev. 39, 305–320 (1986). Metabolic control analysis as the progenitor of metabolic engineering; why the metabolome is expected to be more discriminatory than the transcriptome or proteome.
Savageau, M. Biochemical Systems Analysis: A Study of Function and Design in Molecular Biology (Addison–Wesley, Reading, 1976).
Kitano, H. Systems biology: a brief overview. Science 295, 1662–1664 (2002).
Orchard, S., Zhu, W., Julian, R. J., Hermjakob, H. & Apweiler, R. Further advances in the development of a data interchange standard for proteomics data. Proteomics 3, 2065–2066 (2003).
Orchard, S. et al. Common interchange standards for proteomics data: public availability of tools and schema. Proteomics 4, 490–491 (2004).
Kell, D. B. & King, R. D. On the optimization of classes for the assignment of unidentified reading frames in functional genomics programmes: the need for machine learning. Trends Biotechnol. 18, 93–98 (2000). Why supervised learning methods are more useful than unsupervised ones for functional genomics, and which constraints this entails.
Plumb, R. et al. Metabonomic analysis of mouse urine by liquid-chromatography-time of flight mass spectrometry (LC-TOFMS): detection of strain, diurnal and gender differences. Analyst 128, 819–823 (2003).
Plumb, R. et al. Ultra-performance liquid chromatography coupled to quadrupole- orthogonal time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 18, 2331–2337 (2004). Very high resolution metabolomics using ultra-performance liquid chromatography.
Bailey, J. E. Toward a science of metabolic engineering. Science 252, 1668–1675 (1991).
Stephanopoulos, G. & Sinskey, A. J. Metabolic engineering — methodologies and future prospects. Trends Biotechnol. 11, 392–396 (1993).
Holms, W. H., Hamilton, I. D. & Mousdale, D. Improvements to microbial productivity by analysis of metabolic fluxes. J. Chem. Technol. Biotechnol. 50, 139–141 (1991).
Askenazi, M. et al. Integrating transcriptional and metabolite profiles to direct the engineering of lovastatin-producing fungal strains. Nature Biotechnol. 21, 150–156 (2003).
Reed, J. L. & Palsson, B. Ø. Thirteen years of building constraint-based in silico models of Escherichia coli. J. Bacteriol. 185, 2692–2699 (2003).
Duarte, N. C., Herrgard, M. J. & Palsson, B. O. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 14, 1298–1309 (2004). A genome-scale metabolic model for Saccharomyces cerevisiae.
Kacser, H. The control of enzyme systems in vivo: elasticity analysis of the steady state. Biochem. Soc. Trans. 11, 35–40 (1983).
Pritchard, L. & Kell, D. B. Schemes of flux control in a model of Saccharomyces cerevisiae glycolysis. Eur. J. Biochem. 269, 3894–3904 (2002).
Mendes, P. & Kell, D. B. Non-linear optimization of biochemical pathways: applications to metabolic engineering and parameter estimation. Bioinformatics 14, 869–883 (1998).
van der Greef, J. et al. in Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis (eds Harrigan, G. G. & Goodacre, R.) 171–198 (Kluwer Academic Publishers, Boston, 2003).
McLuhan, M. & Fiore, Q. The Medium is the Massage (Penguin Books, London, 1971).
Koza, J. R. Genetic Programming: on the Programming of Computers by Means of Natural Selection (MIT Press, Cambridge, 1992).
Banzhaf, W., Nordin, P., Keller, R. E. & Francone, F. D. Genetic Programming: An Introduction (Morgan Kaufmann, San Francisco, 1998).
Kell, D. B., Darby, R. M. & Draper, J. Genomic computing: explanatory analysis of plant expression profiling data using machine learning. Plant Physiol. 126, 943–951 (2001).
Kell, D. B. Defence against the flood: a solution to the data mining and predictive modelling challenges of today. Bioinformatics World (part of Scientific Computing News) 1, 16–18 (2002).
Goodacre, R. & Kell, D. B. in Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis (eds Harrigan, G. G. & Goodacre, R.) 239–256 (Kluwer Academic Publishers, Boston, 2003).
Bäck, T., Fogel, D. B. & Michalewicz, Z. (eds) Handbook of Evolutionary Computation (IOP Publishing/Oxford University Press, Oxford, 1997).
Stanislaus, R., Jiang, L., Swartz, M., Arthur, J. & Almeida, J. An XML standard for the dissemination of annotated 2D gel electrophoresis data complemented with mass spectrometry results. BMC Bioinformatics 5 (2004).
Kramer, G., Ruehl, M. & Schafer, R. SpectroML — a markup language for molecular spectrometry data. J. Assoc. Lab. Automat. 6, 76–82 (2001).
Davies, T., Lampen, P., Fiege, M., Richter, T. & Frohlich, T. AnIMLs in the spectroscopic laboratory? Spectroscopy Europe 15, 25–28 (2003).
Shapiro, B. E., Hucka, M., Finney, A. & Doyle, J. MathSBML: a package for manipulating SBML-based biological models. Bioinformatics 20, 2829–2831 (2004).
Acknowledgements
We thank the Biotechnology and Biological Sciences Research Council and the Natural Environment Research Council for financial support. D.B.K. is supported by the Royal Society of Chemistry and the Engineering and Physical Sciences Research Council. We thank N. Bukowski, J. Heald and H. Major for assistance with chromatographic and mass spectrometric analyses, and N. Burton for assistance with yeast microbiology and genetics.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
D.B.K. is a Director of Predictive Solutions Ltd (http://www.abergc.com and http://www.predictivesolutions.co.uk), a company that sells Genetic Programming software.
Related links
Related links
FURTHER INFORMATION
Java Web Services Developer Pack
Glossary
- METABOLOME
-
Nominally all of the small-molecular-weight metabolites in a sample. For practical reasons, this is rarely, if ever, achieved using a single extraction method, and subsets of metabolites ('metabolic profiles') are more typically obtained.
- TRANSCRIPTOME
-
All the mRNA molecules (transcripts) in a sample.
- PROTEOME
-
Nominally all the proteins in a sample. This measurement is not usually achieved because of poor solubilities, especially of membrane proteins, and sometimes protein digests are used, from which peptides representing the proteome are analysed. The concept of the proteome should also include all the post-translational modifications that might occur.
- CE-MS
-
A technique in which metabolites are separated using capillary electrophoresis and then analysed using mass spectrometry. Two different runs are used for cations and anions, whereas neutral molecules can be separated using techniques that give them an effective charge.
- GC-MS
-
A high-resolution analytical technique in which molecules, typically derivatized to enhance their volatility, are separated and then identified using mass spectrometry.
- LC-MS
-
A technique in which metabolites are separated according to their polarity. In metabolomics, reverse-phase chromatography (in which the column is hydrophobic and an organic solvent such as methanol or acetonitrile is used for elution) is most popular, although polar chromatographies are also used.
- METABOLIC FOOTPRINTING
-
A strategy for analysing the properties of cells or tissues by looking in a high-throughout manner at the metabolites that they excrete or fail to take up from their surroundings.
- METABOLIC FINGERPRINTING
-
Classification of samples on the basis of their biological status or origin, using high-throughput methods, usually spectroscopic.
- GENETIC PROGRAMMING
-
A powerful but simple computational technique with which rules are evolved that can be used to solve classification or regression problems, for instance by using the metabolome or metabolic fingerprinting data as the inputs.
- GC-TOF MS
-
A high-resolution analytical technique in which molecules, typically derivatized to enhance their volatility, are separated and then identified using time-of-flight mass spectrometry.
- GC × GC-tof MS
-
Similar to GC-tof MS, but involving two stages of gas-chromatographic separation in which a sample is taken (split) from the first dimension of the separation (typically carried out using a non-polar stationary-phase material) and effecting further ‘orthogonal’ separation (typically using a more polar stationary phase).
- 2DE/MS
-
A proteomics technique in which proteins are separated according to their mass and charge, using two-dimensional gel electrophoresis, and subsequently identified, and sometimes quantified, using mass spectrometric methods.
Rights and permissions
About this article
Cite this article
Kell, D., Brown, M., Davey, H. et al. Metabolic footprinting and systems biology: the medium is the message. Nat Rev Microbiol 3, 557–565 (2005). https://doi.org/10.1038/nrmicro1177
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1177
This article is cited by
-
Resourcefulness of propylprodigiosin isolated from Brevundimonas olei strain RUN-D1
AMB Express (2023)
-
Vitreous metabolomic signatures of pathological myopia with complications
Eye (2023)
-
Chemical Profiling of Volatile Bioactives in Luisia tenuifolia Blume Successive Extracts by GC–MS Analysis
Applied Biochemistry and Biotechnology (2022)
-
Comparative analysis of gender-specific metabolomic discriminators in two species of seabuckthorn (Hippophae rhamnoides L. and Hippophae salicifolia D.Don) from different regions of Indian Himalayas
Plant Physiology Reports (2022)