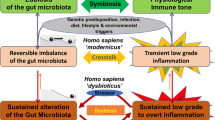
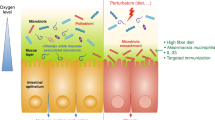
Abstract
The composition of the intestinal microbiota varies among individuals and throughout development, and is dependent on host and environmental factors. However, although the microbiota is constantly exposed to environmental challenges, its composition and function in an individual are stable against perturbations, as microbial communities are resilient and resistant to change. The maintenance of a beneficial microbiota requires a homeostatic equilibrium within microbial communities, and also between the microorganisms and the intestinal interface of the host. The resilience of the healthy microbiota protects us from dysbiosis-related diseases, such as inflammatory bowel disease (IBD) or metabolic disorder. By contrast, a resilient dysbiotic microbiota may cause disease. In this Opinion article, we propose that microbial resilience has a key role in health and disease. We will discuss the concepts and mechanisms of microbial resilience against dietary, antibiotic or bacteriotherapy-induced perturbations and the implications for human health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Faith, J. J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Caporaso, J. G. et al. Moving pictures of the human microbiome. Genome Biol. 12, R50 (2011).
Walker, B., Holling, C. S., Carpenter, S. R. & Kinzig, A. Resilience, adaptability and transformability in social–ecological systems. Ecol. Soc. 9, 5 (2004).
Moya, A. & Ferrer, M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 24, 402–413 (2016).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Greenhalgh, K., Meyer, K. M., Aagaard, K. M. & Wilmes, P. The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ. Microbiol. 18, 2103–2116 (2016).
Chassaing, B. et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 (2015).
Suez, J. et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 514, 181–186 (2014).
Henle, J. in Pathologische Untersuchungen 1–82 (Hirschwald, 1840).
Koch, R. Die Ätiologie der Tuberkulose [German]. Berliner Klinische Wochenschrift 19, 428–445 (1882).
Singh, V. P., Proctor, S. D. & Willing, B. P. Koch's postulates, microbial dysbiosis and inflammatory bowel disease. Clin. Microbiol. Infect. 22, 594–599 (2016).
Shade, A. et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3, 417 (2012).
Bender, E. A., Case, T. J. & Gilpin, M. E. Perturbation experiments in community ecology: theory and practice. Ecology 65, 1–13 (1984).
Oliver, T. H. et al. Biodiversity and resilience of ecosystem functions. Trends Ecol. Evol. 30, 673–684 (2015).
Gunderson, L. H. Ecological resilience — in theory and application. Annu. Rev. Ecol. Syst. 31, 425–439 (2000).
Holling, C. S. & Gunderson, L. in Panarchy: Understanding Transformations in Human and Natural Systems 25–62 (Island Press, 2002).
Holling, C. S. Resilience and stability of ecological systems. Annu. Rev. Ecol. Syst. 4, 1–23 (1973).
Sommer, F. & Backhed, F. Know your neighbor: microbiota and host epithelial cells interact locally to control intestinal function and physiology. Bioessays 38, 455–464 (2016).
Wang, J. et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc. Natl Acad. Sci. USA 111, E2703–E2710 (2014).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
McNaughton, S. J. Diversity and stability of ecological communities: a comment on the role of empiricism in ecology. Am. Nat. 111, 515–525 (1977).
Naeem, S. & Li, S. Biodiversity enhances ecosystem reliability. Nature 390, 507–509 (1997).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
Ley, R. E. et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA 102, 11070–11075 (2005).
Furet, J. P. et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes 59, 3049–3057 (2010).
Manichanh, C. et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211 (2006).
Willing, B. P. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854.e1 (2010).
Ott, S. J. et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53, 685–693 (2004).
Lepage, P. et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141, 227–236 (2011).
Chang, J. Y. et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197, 435–438 (2008).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
Moeller, A. H. et al. Chimpanzees and humans harbour compositionally similar gut enterotypes. Nat. Commun. 3, 1179 (2012).
Horst, K. et al. Risk stratification by injury distribution in polytrauma patients — does the clavicular fracture play a role? Patient Saf. Surg. 7, 23 (2013).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
Vieira-Silva, S. et al. Species–function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 1, 16088 (2016).
Schreiber, S., Rosenstiel, P., Albrecht, M., Hampe, J. & Krawczak, M. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat. Rev. Genet. 6, 376–388 (2005).
Hsiao, A. et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426 (2014).
Schwab, C. et al. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J. 8, 1101–1114 (2014).
Buffie, C. G. et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80, 62–73 (2012).
Nemergut, D. R. et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356 (2013).
Seedorf, H. et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 159, 253–266 (2014).
Salzman, N. H. et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–83 (2010).
Sommer, F. et al. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS ONE 9, e85254 (2014).
Johansson, M. E. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA 105, 15064–15069 (2008).
Hooper, L. V., Littman, D. R. & Macpherson, A. J. Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012).
Wehkamp, J. et al. Reduced Paneth cell α-defensins in ileal Crohn's disease. Proc. Natl Acad. Sci. USA 102, 18129–18134 (2005).
Mathewson, N. D. et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol. 17, 505–513 (2016).
Nigro, G., Rossi, R., Commere, P. H., Jay, P. & Sansonetti, P. J. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe 15, 792–798 (2014).
Lindemans, C. A. et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015).
Hadizadeh, F. et al. Stool frequency is associated with gut microbiota composition. Gut 66, 559–560 (2016).
Falony, G. et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016).
Alam, A. et al. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat. Microbiol. 1, 15021 (2016).
Pedron, T. et al. A crypt-specific core microbiota resides in the mouse colon. mBio 3, e00116-12 (2012).
Gillilland, M. G. III et al. Ecological succession of bacterial communities during conventionalization of germ-free mice. Appl. Environ. Microbiol. 78, 2359–2366 (2012).
Wier, A. M. et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc. Natl Acad. Sci. USA 107, 2259–2264 (2010).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Thaiss, C. A. et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529 (2014).
Thaiss, C. A. et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167, 1495–1510.e12 (2016).
Faust, K. & Raes, J. Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550 (2012).
Jones, S. E. & Lennon, J. T. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl Acad. Sci. USA 107, 5881–5886 (2010).
Pedros-Alio, C. Marine microbial diversity: can it be determined? Trends Microbiol. 14, 257–263 (2006).
Lennon, J. T. & Jones, S. E. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130 (2011).
Tait, K. & Sutherland, I. W. Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. J. Appl. Microbiol. 93, 345–352 (2002).
LaSarre, B. & Federle, M. J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111 (2013).
Tan, C. H. et al. Community quorum sensing signalling and quenching: microbial granular biofilm assembly. NPJ Biofilms Microbiomes 1, 15006 (2015).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Sommer, F. & Backhed, F. The gut microbiota — masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238 (2013).
Rehman, A. et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut 60, 1354–1362 (2011).
Rakoff-Nahoum, S. et al. Analysis of gene–environment interactions in postnatal development of the mammalian intestine. Proc. Natl Acad. Sci. USA 112, 1929–1936 (2015).
Koenig, J. E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4578–4585 (2011).
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Russell, S. L. et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447 (2012).
Dominguez-Bello, M. G. et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 22, 250–253 (2016).
Subramanian, S. et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014).
Dethlefsen, L. & Relman, D. A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4554–4561 (2011).
Heinsen, F. A. et al. Dynamic changes of the luminal and mucosa-associated gut microbiota during and after antibiotic therapy with paromomycin. Gut Microbes 6, 243–254 (2015).
Jernberg, C., Lofmark, S., Edlund, C. & Jansson, J. K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66 (2007).
Lindner, C. et al. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat. Immunol. 16, 880–888 (2015).
Schumann, A. et al. Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol. Genomics 23, 235–245 (2005).
Biedermann, L. & Rogler, G. The intestinal microbiota: its role in health and disease. Eur. J. Pediatr. 174, 151–167 (2015).
Theriot, C. M., Bowman, A. A. & Young, V. B. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere http://dx.doi.org/10.1128/mSphere.00045-15 (2016).
Risnes, K. R., Belanger, K., Murk, W. & Bracken, M. B. Antibiotic exposure by 6 months and asthma and allergy at 6 years: findings in a cohort of 1,401 US children. Am. J. Epidemiol. 173, 310–318 (2011).
Shaw, S. Y., Blanchard, J. F. & Bernstein, C. N. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am. J. Gastroenterol. 105, 2687–2692 (2010).
Kronman, M. P., Zaoutis, T. E., Haynes, K., Feng, R. & Coffin, S. E. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics 130, e794–e803 (2012).
Azad, M. B., Bridgman, S. L., Becker, A. B. & Kozyrskyj, A. L. Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int. J. Obes. (Lond.) 38, 1290–1298 (2014).
Boursi, B., Mamtani, R., Haynes, K. & Yang, Y. X. The effect of past antibiotic exposure on diabetes risk. Eur. J. Endocrinol. 172, 639–648 (2015).
Blaser, M. J. Antibiotic use and its consequences for the normal microbiome. Science 352, 544–545 (2016).
Vrieze, A. et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916.e7 (2012).
Bäckhed, F. et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl Acad. Sci. USA 101, 15718–15723 (2004).
Couturier-Maillard, A. et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123, 700–711 (2013).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010).
Hamilton, M. J., Weingarden, A. R., Unno, T., Khoruts, A. & Sadowsky, M. J. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes 4, 125–135 (2013).
Li, S. S. et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 352, 586–589 (2016).
Manichanh, C. et al. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 20, 1411–1419 (2010).
Fuentes, S. et al. Reset of a critically disturbed microbial ecosystem: faecal transplant in recurrent Clostridium difficile infection. ISME J. 8, 1621–1633 (2014).
Grinspan, A. M. & Kelly, C. R. Fecal microbiota transplantation for ulcerative colitis: not just yet. Gastroenterology 149, 15–18 (2015).
Eiseman, B., Silen, W., Bascom, G. S. & Kauvar, A. J. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery 44, 854–859 (1958).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
Broecker, F., Klumpp, J. & Moelling, K. Long-term microbiota and virome in a Zurich patient after fecal transplantation against Clostridium difficile infection. Ann. NY Acad. Sci. 1372, 29–41 (2016).
Vermeire, S. et al. Donor species richness determines faecal microbiota transplantation success in inflammatory bowel disease. J. Crohns Colitis 10, 387–394 (2016).
Colman, R. J. & Rubin, D. T. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J. Crohns Colitis 8, 1569–1581 (2014).
Moayyedi, P. et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109.e6 (2015).
Rossen, N. G. et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 149, 110–118.e4 (2015).
Ott, S. J. et al. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 152, 799–811.e7 (2016).
Rykiel, E. J. Towards a definition of ecological disturbance. Aust. J. Ecol. 10, 361–365 (1985).
Worm, B. & Duffy, J. E. Biodiversity, productivity and stability in real food webs. Trends Ecol. Evol. 18, 628–632 (2003).
Relman, D. A. The human microbiome: ecosystem resilience and health. Nutr. Rev. 70, S2–S9 (2012).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
David, L. A. et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014).
Costello, E. K., Stagaman, K., Dethlefsen, L., Bohannan, B. J. & Relman, D. A. The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262 (2012).
Tamboli, C. P., Neut, C., Desreumaux, P. & Colombel, J. F. Dysbiosis in inflammatory bowel disease. Gut 53, 1–4 (2004).
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; grants CRC1182 C2 and CRC877 B9), the Federal Ministry of Education and Research as part of the e:Med framework ('sysINFLAME'; grant 01ZX1306), the Cluster of Excellence 'Inflammation at Interfaces' (grant ExC 306) and SYSCID (a systems medicine approach to chronic inflammatory diseases) in the European Union's Horizon 2020 research and innovation programme under grant agreement No 733100. The authors express that this article represents an opinionated perspective rather than a systematic review. The authors apologize to those researchers whose important contribution to the field could not be cited owing to space constraints.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Sommer, F., Anderson, J., Bharti, R. et al. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol 15, 630–638 (2017). https://doi.org/10.1038/nrmicro.2017.58
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2017.58
This article is cited by
-
The role of gut microbiota in human metabolism and inflammatory diseases: a focus on elderly individuals
Annals of Microbiology (2024)
-
Effects of removing in-feed antibiotics and zinc oxide on the taxonomy and functionality of the microbiota in post weaning pigs
Animal Microbiome (2024)
-
Exploring the potential effects of forest urbanization on the interplay between small mammal communities and their gut microbiota
Animal Microbiome (2024)
-
Unlocking the secrets: exploring the influence of the aryl hydrocarbon receptor and microbiome on cancer development
Cellular & Molecular Biology Letters (2024)
-
Impact of the diet in the gut microbiota after an inter-species microbial transplantation in fish
Scientific Reports (2024)