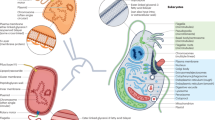
Key Points
-
Genome fragments are sometimes transferred from bacteria to eukaryotes via horizontal gene transfer (HGT).
-
When these DNA fragments contain genes, these genes can retain their functionality in some cases.
-
If these bacterial sequences are maintained for long periods of time, they can acquire eukaryotic features such as introns.
-
If the eukaryotic recipient retains a stable bacterial endosymbiont, these HGT events can compensate for genome reduction in the endosymbiont.
-
These HGT events can also enable the eukaryotic recipient to protect itself from other organisms, survive in new environments or use new food sources.
-
Further study of neglected eukaryotic groups will help to clarify the frequency of bacteria–eukaryote HGT.
Abstract
Bacteria influence eukaryotic biology as parasitic, commensal or beneficial symbionts. Aside from these organismal interactions, bacteria have also been important sources of new genetic sequences through horizontal gene transfer (HGT) for eukaryotes. In this Review, we focus on gene transfers from bacteria to eukaryotes, discuss how horizontally transferred genes become functional and explore what functions are endowed upon a broad diversity of eukaryotes by genes derived from bacteria. We classify HGT events into two broad types: those that maintain pre-existing functions and those that provide the recipient with new functionality, including altered host nutrition, protection and adaptation to extreme environments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Koonin, E. V., Makarova, K. S. & Aravind, L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55, 709–742 (2001).
Gogarten, J. P. & Townsend, J. P. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3, 679–687 (2005).
Thomas, C. M. & Nielsen, K. M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721 (2005).
Soucy, S. M., Huang, J. & Gogarten, J. P. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472–482 (2015).
Koonin, E. V. Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Res. 5, 1805 (2016).
Wagner, A. et al. Mechanisms of gene flow in archaea. Nat. Rev. Microbiol. 15, 492–501 (2017).
Syvanen, M. Evolutionary implications of horizontal gene transfer. Annu. Rev. Genet. 46, 341–358 (2012).
Doolittle, W. F. et al. What is the tree of life? PLOS Genet. 12, e1005912 (2016).
Philippe, H. & Douady, C. J. Horizontal gene transfer and phylogenetics. Curr. Opin. Microbiol. 6, 498–505 (2003).
Timmis, J. N., Ayliffe, M. A., Huang, C. Y. & Martin, W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (2004).
Keeling, P. J. & Palmer, J. D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618 (2008).
Alsmark, C. et al. Patterns of prokaryotic lateral gene transfers affecting parasitic microbial eukaryotes. Genome Biol. 14, R19 (2013). This is a systematic reanalysis of 13 parasitic microbial eukaryotes that shows that the majority of HGT events in these species involve genes related to amino acid and sugar metabolism and that putative donors of these genes are significantly enriched in bacterial groups sharing the same habitats.
Schönknecht, G., Weber, A. P. M. & Lercher, M. J. Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. Bioessays 36, 9–20 (2014).
Lacroix, B. & Citovsky, V. Transfer of DNA from bacteria to eukaryotes. MBio 7, e00863–e00816 (2016).
Robinson, K. M., Sieber, K. B. & Dunning Hotopp, J. C. A review of bacteria-animal lateral gene transfer may inform our understanding of diseases like cancer. PLoS Genet. 9, e1003877 (2013).
Boto, L. Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc. Biol. Sci. 281, 20132450 (2014).
Wybouw, N., Pauchet, Y., Heckel, D. G. & Van Leeuwen, T. Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol. Evol. 8, 1785–1801 (2016).
Salzberg, S. L. et al. Microbial genes in the human genome: lateral transfer or gene loss? Science 292, 1903–1906 (2001).
Salzberg, S. L. Horizontal gene transfer is not a hallmark of the human genome. Genome Biol. 18, 85 (2017).
Crisp, A. et al. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 16, 50 (2015).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Stanhope, M. J. et al. Phylogenetic analyses do not support horizontal gene transfers from bacteria to vertebrates. Nature 411, 940–944 (2001).
Arakawa, K. No evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc. Natl Acad. Sci. USA 113, E3057 (2016).
Koutsovoulos, G. et al. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc. Natl Acad. Sci. USA 113, 5053–5058 (2016).
Nikoh, N. et al. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. Plos Genet. 6, e1000827 (2010).
Moran, Y., Fredman, D., Szczesny, P., Grynberg, M. & Technau, U. Recurrent horizontal transfer of bacterial toxin genes to eukaryotes. Mol. Biol. Evol. 29, 2223–2230 (2012). Bacteria use pore-forming toxins to lyse cellular membranes, but this article reveals several independent transfers of the genes for these toxins to eukaryotic genomes and discusses the possible role of those genes in defence and predation of multicellular eukaryotes.
Flot, J. et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500, 453–457 (2014).
Husnik, F. et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153, 1567–1578 (2013).
Wu, B. et al. Interdomain lateral gene transfer of an essential ferrochelatase gene in human parasitic nematodes. Proc. Natl Acad. Sci. USA 110, 7748–7753 (2013).
Chou, S. et al. Transferred interbacterial antagonism genes augment eukaryotic innate immune function. Nature 518, 98–101 (2014). This is an important study that shows multiple independent HGT events of genes encoding antibacterial amidases across the tree of life and experimentally verifies their function.
Metcalf, J. A., Funkhouser-Jones, L. J., Brileya, K., Reysenbach, A.-L. & Bordenstein, S. R. Antibacterial gene transfer across the tree of life. eLife 3, e04266 (2014). This article shows and experimentally verifies HGT events for genes encoding bacterial cell wall degradation enzymes (lysozymes) occurring in parallel across the tree of life.
Sloan, D. B. et al. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol. Biol. Evol. 31, 857–871 (2014).
Wybouw, N. et al. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. eLife 3, e02365 (2014). This is an elegant analysis of HGT events of a cysteine synthase family found in several lineages of herbivorous arthropods. Functional expression analysis suggests that the enzymes can be used for both cysteine biosynthesis and cyanide detoxification, but enzyme kinetics suggest their main function is cyanide detoxification.
Luan, J.-B. et al. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol. Evol. 7, 2635–2647 (2015).
Baltrus, D. A. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495 (2013).
Koutsovoulos, G. et al. Palaeosymbiosis revealed by genomic fossils of Wolbachia in a strongyloidean nematode. PLoS Genet. 10, e1004397 (2014).
Choi, J. Y., Bubnell, J. E. & Aquadro, C. F. Population genomics of infectious and integrated Wolbachia pipientis genomes in Drosophila ananassae. Genome Biol. Evol. 7, 2362–2382 (2015).
Derelle, E. et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl Acad. Sci. USA 103, 11647–11652 (2006).
Eyres, I. et al. Horizontal gene transfer in bdelloid rotifers is ancient, ongoing and more frequent in species from desiccating habitats. BMC Biol. 13, 90 (2015).
Ku, C. et al. Endosymbiotic gene transfer from prokaryotic pangenomes: inherited chimerism in eukaryotes. Proc. Natl Acad. Sci. USA 112, 10139–10146 (2015).
Ku, C. et al. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524, 427–432 (2015).
Katz, L. A. Recent events dominate interdomain lateral gene transfers between prokaryotes and eukaryotes and, with the exception of endosymbiotic gene transfers, few ancient transfer events persist. Phil. Trans. R. Soc. B Biol. Sci. 370, 20140324 (2015).
Brown, J. R. Ancient horizontal gene transfer. Nat. Rev. Genet. 4, 121–132 (2003).
Nowack, E. C. M. et al. Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proc. Natl Acad. Sci. USA 113, 12214–12219 (2016). Paulinella chromatophora is an amoeba that contains a photosynthetic symbiont. Genomic and transcriptomic data enabled the authors to untangle how many bacterial genes in the genome of P. chromatophora came from phylogenetic sources other than the current symbiont and suggest that HGT from diverse bacteria compensates for gene loss in the symbiont.
Xu, F. et al. On the reversibility of parasitism: adaptation to a free-living lifestyle via gene acquisitions in the diplomonad Trepomonas sp. PC1. BMC Biol. 14, 62 (2016).
Paganini, J. et al. Contribution of lateral gene transfers to the genome composition and parasitic ability of root-knot nematodes. PLoS ONE 7, e50875 (2012).
Schönknecht, G. et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339, 1207–1210 (2013). This study describes the important role that HGT from bacteria has had in enabling the red alga Galdieria to adapt to hot, toxic and acidic environments.
Qiu, H. et al. Adaptation through horizontal gene transfer in the cryptoendolithic red alga Galdieria phlegrea. Curr. Biol. 23, R865–R866 (2013).
Savory, F., Leonard, G. & Richards, T. A. The role of horizontal gene transfer in the evolution of the oomycetes. PLoS Pathog. 11, e1004805 (2015). This study evaluates extensive HGT from bacteria and fungi that assisted oomycetes in becoming successful plant parasites.
Eme, L., Gentekaki, E., Curtis, B., Archibald, J. M. & Roger, A. J. Lateral gene transfer in the adaptation of the anaerobic parasite Blastocystis to the gut. Curr. Biol. 27, 807–820 (2017). This rigorous phylogeny-based analysis of HGT in Blastocystis spp. shows that 2.5% of their genes have been fairly recently acquired from bacteria, archaea and eukaryotes. In particular, the authors report genes involved in carbohydrate scavenging and metabolism, anaerobic amino acid and nitrogen metabolism, oxygen-stress resistance and pH homeostasis. These HGT events are hypothesized to assist Blastocystis spp. in adaptation to the gut environment they inhabit.
Husnik, F. & McCutcheon, J. P. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc. Natl Acad. Sci. USA 113, E5416–E5424 (2016). One common argument against the idea of HGT from diverse bacteria compensating for symbiont or organelle gene loss is that complex pathways are impossible to build in a gene-by-gene fashion. This study shows how complex and mosaic pathways can be built sequentially with HGT events from diverse bacteria and that pre-existing HGT events can remain stable in genomes in the face of extensive symbiont turnover.
Boschetti, C. et al. Biochemical diversification through foreign gene expression in bdelloid rotifers. PLoS Genet. 8, e1003035 (2012).
Marcet-Houben, M. & Gabaldón, T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 26, 5–8 (2010).
Ford Doolittle, W. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14, 307–311 (1998).
Gluck-Thaler, E. & Slot, J. C. Dimensions of horizontal gene transfer in eukaryotic microbial pathogens. PLoS Pathog. 11, e1005156 (2015).
Huang, J. Horizontal gene transfer in eukaryotes: the weak-link model. Bioessays 35, 868–875 (2013). This study presents the weak-link model, which posits that foreign genes enter eukaryotic genomes at unprotected stages of their development; the model thus expands previous models for multicellular sexual eukaryotes.
Dunning Hotopp, J. C. Horizontal gene transfer between bacteria and animals. Trends Genet. 27, 157–163 (2011).
Brelsfoard, C. et al. Presence of extensive Wolbachia symbiont insertions discovered in the genome of its host Glossina morsitans morsitans. PLoS Negl. Trop. Dis. 8, e2728 (2014).
Klasson, L. et al. Extensive duplication of the Wolbachia DNA in chromosome four of Drosophila ananassae. BMC Genomics 15, 1097 (2014).
Lieber, M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
Hao, W., Richardson, A. O., Zheng, Y. & Palmer, J. D. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc. Natl Acad. Sci. USA 107, 21576–21581 (2010).
Dunning Hotopp, J. C. et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 (2007).
Funkhouser-Jones, L. J. et al. Wolbachia co-infection in a hybrid zone: discovery of horizontal gene transfers from two Wolbachia supergroups into an animal genome. PeerJ 3, e1479 (2015).
Leclercq, S. et al. Birth of a W sex chromosome by horizontal transfer of Wolbachia bacterial symbiont genome. Proc. Natl Acad. Sci. USA 113, 15036–15041 (2016).
Kumar, N. et al. Efficient subtraction of insect rRNA prior to transcriptome analysis of Wolbachia-Drosophila lateral gene transfer. BMC Res. Notes 5, 230 (2012).
Ricchetti, M., Tekaia, F. & Dujon, B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2, E273 (2004).
Ricchetti, M., Fairhead, C. & Dujon, B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature 402, 96–100 (1999).
Tsuji, J., Frith, M. C., Tomii, K. & Horton, P. Mammalian NUMT insertion is non-random. Nucleic Acids Res. 40, 9073–9088 (2012).
McNulty, S. N. et al. Endosymbiont DNA in endobacteria-free filarial nematodes indicates ancient horizontal genetic transfer. PLoS ONE 5, e11029 (2010).
Acuna, R. et al. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc. Natl Acad. Sci. USA 109, 4197–4202 (2012).
Pauchet, Y. & Heckel, D. G. The genome of the mustard leaf beetle encodes two active xylanases originally acquired from bacteria through horizontal gene transfer. Proc. Biol. Sci. 280, 20131021 (2013).
Gladyshev, E. A., Meselson, M. & Arkhipova, I. R. Massive horizontal gene transfer in bdelloid rotifers. Science 320, 1210–1213 (2008). This is one of the first studies to describe massive HGT to an animal genome with details on gene localization, structure and function.
Sun, B. F. et al. Multiple ancient horizontal gene transfers and duplications in lepidopteran species. Insect Mol. Biol. 22, 72–87 (2013).
Le Hir, H., Nott, A. & Moore, M. J. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28, 215–220 (2003).
Craig, J. P., Bekal, S., Niblack, T., Domier, L. & Lambert, K. N. Evidence for horizontally transferred genes involved in the biosynthesis of vitamin B1, B5, and B7 in Heterodera glycines. J. Nematol. 41, 281–290 (2009).
Bowler, C. et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244 (2008).
Danchin, E. G. J. et al. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc. Natl Acad. Sci. USA 107, 17651–17656 (2010).
Andersson, J. O. et al. Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62, 1182–1197 (2005).
Archibald, J. M. Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 25, R911–R921 (2015).
Gray, M. W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 4, a011403 (2012).
Koonin, E. V. & Yutin, N. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb. Perspect. Biol. 6, a016188 (2014).
Ponce-Toledo, R. I. et al. An early-branching freshwater cyanobacterium at the origin of plastids. Curr. Biol. 27, 386–391 (2017).
Wang, Z. & Wu, M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 5, 7949 (2015).
Gray, M. W. Mosaic nature of the mitochondrial proteome: implications for the origin and evolution of mitochondria. Proc. Natl Acad. Sci. USA 112, 10133–10138 (2015).
Huynen, M. A., Duarte, I. & Szklarczyk, R. Loss, replacement and gain of proteins at the origin of the mitochondria. Biochim. Biophys. Acta 1827, 224–231 (2013).
Gabaldón, T. & Huynen, M. A. From endosymbiont to host-controlled organelle: the hijacking of mitochondrial protein synthesis and metabolism. PLoS Comput. Biol. 3, e219 (2007).
Esser, C. et al. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21, 1643–1660 (2004).
Thiergart, T., Landan, G., Schenk, M., Dagan, T. & Martin, W. F. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol. Evol. 4, 466–485 (2012).
Kurland, C. G. & Andersson, S. G. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 64, 786–820 (2000).
Suzuki, K. & Miyagishima, S. Eukaryotic and eubacterial contributions to the establishment of plastid proteome estimated by large-scale phylogenetic analyses. Mol. Biol. Evol. 27, 581–590 (2010).
Zaremba-Niedzwiedzka, K. et al. Asgard arcahea illuminates the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
Pittis, A. A. & Gabaldón, T. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531, 101–104 (2016).
Koonin, E. V. Archaeal ancestors of eukaryotes: not so elusive any more. BMC Biol. 13, 84 (2015).
Spang, A. et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015).
Delaye, L., Valadez-Cano, C. & Pérez-Zamorano, B. How really ancient is Paulinella chromatophora? PLoS Curr. http://dx.doi.org/10.1371/currents.tol.e68a099364bb1a1e129a17b4e06b0c6b (2016).
Klein, C. C. et al. Biosynthesis of vitamins and cofactors in bacterium-harbouring trypanosomatids depends on the symbiotic association as revealed by genomic analyses. PLoS ONE 8, e79786 (2013).
Alves, J. M. P. et al. Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol. Biol. 13, 190 (2013).
Larkum, A. W. D., Lockhart, P. J. & Howe, C. J. Shopping for plastids. Trends Plant Sci. 12, 189–195 (2007).
Gray, M. W. The pre-endosymbiont hypothesis: a new perspective on the origin and evolution of mitochondria. Cold Spring Harb. Perspect. Biol. 6, a016097 (2014).
Qiu, H. et al. Assessing the bacterial contribution to the plastid proteome. Trends Plant Sci. 18, 680–687 (2013).
Karnkowska, A. et al. A eukaryote without a mitochondrial organelle. Curr. Biol. 26, 1274–1284 (2016).
Embley, T. M. & Martin, W. Eukaryotic evolution, changes and challenges. Nature 440, 623–630 (2006).
Daimon, T. et al. β-fructofuranosidase genes of the silkworm, Bombyx mori: insights into enzymatic adaptation of B. mori to toxic alkaloids in mulberry latex. J. Biol. Chem. 283, 15271–15279 (2008).
Ambrose, K. V., Koppenhöfer, A. M. & Belanger, F. C. Horizontal gene transfer of a bacterial insect toxin gene into the Epichloë fungal symbionts of grasses. Sci. Rep. 4, 5562 (2014).
Daborn, P. J. et al. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc. Natl Acad. Sci. USA 99, 10742–10747 (2002).
Ricard, G. et al. Horizontal gene transfer from bacteria to rumen ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics 7, 22 (2006).
Richards, T. A., Leonard, G., Soanes, D. M. & Talbot, N. J. Gene transfer into the fungi. Fungal Biol. Rev. 25, 98–110 (2011).
Danchin, E. G. J. & Rosso, M.-N. Lateral gene transfers have polished animal genomes: lessons from nematodes. Front. Cell. Infect. Microbiol. 2, 27 (2012).
Schuster, L. N. & Sommer, R. J. Expressional and functional variation of horizontally acquired cellulases in the nematode Pristionchus pacificus. Gene 506, 274–282 (2012).
Sasakura, Y. et al. Transcriptional regulation of a horizontally transferred gene from bacterium to chordate. Proc. Biol. Sci. 283, 20161712 (2016).
Huang, J. et al. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 5, R88 (2004).
Marchetti, A. et al. Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature 457, 467–470 (2009).
Allen, A. E. et al. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl Acad. Sci. USA 105, 10438–10443 (2008).
Eichinger, L. et al. The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 (2005).
Andersson, J. O. Evolution of patchily distributed proteins shared between eukaryotes and prokaryotes: Dictyostelium as a case study. J. Mol. Microbiol. Biotechnol. 20, 83–95 (2011).
Worden, A. Z. et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 375, 268–272 (2009).
de Koning, A. P., Brinkman, F. S. L., Jones, S. J. M. & Keeling, P. J. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol. Biol. Evol. 17, 1769–1773 (2000).
Sun, G. & Huang, J. Horizontally acquired DAP pathway as a unit of self-regulation. J. Evol. Biol. 24, 587–595 (2011).
Craig, J. P. et al. Analysis of a horizontally transferred pathway involved in vitamin B6 biosynthesis from the soybean cyst nematode Heterodera glycines. Mol. Biol. Evol. 25, 2085–2098 (2008).
Mock, T. et al. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541, 536–540 (2017).
Raymond, J. A. et al. Possible role of horizontal gene transfer in the colonization of sea ice by algae. PLoS ONE 7, e35968 (2012).
Blanc, G. et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 13, R39 (2012).
Harding, T., Roger, A. J. & Simpson, A. G. B. Adaptations to high salt in a halophilic protist: differential expression and gene acquisitions through duplications and gene transfers. Front. Microbiol. 8, 944 (2017).
Slamovits, C. H. & Keeling, P. J. Class II photolyase in a microsporidian intracellular parasite. J. Mol. Biol. 341, 713–721 (2004).
Fast, N. M., Law, J. S., Williams, B. A. P. & Keeling, P. J. Bacterial catalase in the microsporidian Nosema locustae: implications for microsporidian metabolism and genome evolution. Eukaryot. Cell 2, 1069–1075 (2003).
Dunning Hotopp, J. C. et al. Biology wars: the eukaryotes strike back. Cell Host Microbe 16, 701–703 (2014).
Ioannidis, P. et al. Rapid transcriptome sequencing of an invasive pest, the brown marmorated stink bug Halyomorpha halys. BMC Genomics 15, 738 (2014).
Yue, J., Hu, X., Sun, H., Yang, Y. & Huang, J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat. Commun. 3, 1152 (2012).
Hirano, T. et al. Moss chloroplasts are surrounded by a peptidoglycan wall containing D-amino acids. Plant Cell 28, 1521–1532 (2016).
Machida, M. et al. Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proc. Natl Acad. Sci. USA 103, 6753–6758 (2006).
Garcia, M. et al. An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J. 53, 924–934 (2008).
Royet, J., Gupta, D. & Dziarski, R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat. Rev. Immunol. 11, 837–851 (2011).
Nyholm, S. V. & Graf, J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 10, 815–827 (2012).
Wang, J. W., Wu, Y. N., Yang, G. X. & Aksoy, S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl Acad. Sci. USA 106, 12133–12138 (2009).
DiSalvo, S. et al. Burkholderia bacteria infectiously induce the proto-farming symbiosis of Dictyostelium amoebae and food bacteria. Proc. Natl Acad. Sci. USA 112, E5029–E5037 (2015).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Boschetti, C. et al. Foreign genes and novel hydrophilic protein genes participate in the desiccation response of the bdelloid rotifer Adineta ricciae. J. Exp. Biol. 214, 59–68 (2011).
Wheeler, D., Redding, A. J. & Werren, J. H. Characterization of an ancient lepidopteran lateral gene transfer. PLoS ONE 8, e59262 (2013).
Philippe, H. et al. Pitfalls in supermatrix phylogenomics. Eur. J. Taxon. 283, 1–25 (2017).
Shimodaira, H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002).
Richards, T. A. et al. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl Acad. Sci. USA 108, 15258–15263 (2011).
Stairs, C. W., Roger, A. J. & Hampl, V. Eukaryotic pyruvate formate lyase and its activating enzyme were acquired laterally from a firmicute. Mol. Biol. Evol. 28, 2087–2099 (2011).
Stechmann, A. et al. The glycolytic pathway of Trimastix pyriformis is an evolutionary mosaic. BMC Evol. Biol. 6, 101 (2006).
Boxma, B. et al. The [FeFe] hydrogenase of Nyctotherus ovalis has a chimeric origin. BMC Evol. Biol. 7, 230 (2007).
Müller, M., Lee, J. A., Gordon, P., Gaasterland, T. & Sensen, C. W. Presence of prokaryotic and eukaryotic species in all subgroups of the PPi-dependent group II phosphofructokinase protein family. J. Bacteriol. 183, 6714–6716 (2001).
Nixon, J. E. J. et al. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot. Cell 1, 181–190 (2002).
Sateriale, A. & Striepen, B. Beg, borrow and steal: three aspects of horizontal gene transfer in the protozoan parasite, Cryptosporidium parvum. PLoS Pathog. 12, e1005429 (2016).
Gojkovic, Z. et al. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol. Genet. Genomics 271, 387–393 (2004).
Tsaousis, A. D. et al. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 453, 553–556 (2008).
Alexander, W. G., Wisecaver, J. H., Rokas, A. & Hittinger, C. T. Horizontally acquired genes in early-diverging pathogenic fungi enable the use of host nucleosides and nucleotides. Proc. Natl Acad. Sci. USA 113, 4116–4121 (2016). This comprehensive analysis of HGT events in Microsporidia and Cryptomycota fungi detects dozens of novel HGT candidates, including parallel acquisitions that enable these pathogenic fungi to scavenge nucleosides and nucleotides from their hosts.
Acknowledgements
F.H. was supported by the Fulbright Commission and a fellowship from the European Molecular Biology Organization (EMBO; ALTF 1260–2016) while writing this Review. J.P.M. was supported by grants from the US National Science Foundation (IOS-1256680 and IOS-1553529), the US National Aeronautics and Space Administration (NASA) Astrobiology Institute (NNA15BB04A) and the Gordon and Betty Moore Foundation (GBMF5602).
Author information
Authors and Affiliations
Contributions
F.H. and J.P.M. researched data for the article, made substantial contributions to discussions of the content, wrote the article and reviewed and/or edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Supplementary information
Supplementary information S1 (table)
HGT events from bacteria to eukaryotes. (XLSX 33 kb)
Glossary
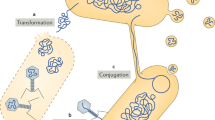
- Horizontal gene transfer
-
(HGT). Movement of genetic material between organisms (also called lateral gene transfer) via non-vertical (not parent-to-offspring) transmission.
- Germ line
-
The specialized cellular lineage in multicellular sexual organisms that is used to pass on genetic material to the progeny.
- Endosymbionts
-
Organisms living within the body or cells of another organism.
- Long-branch attraction
-
A phylogenetic artefact that causes distantly related lineages (often on long branches) to be incorrectly inferred as closely related in phylogenetic trees.
- Conjugation
-
Gene transfer from a donor to a recipient by direct cell-to-cell contact, such as plasmid transfer between two bacterial cells.
- Transduction
-
Gene transfer carried out by a virus, such as a bacteriophage, transferring DNA from one bacterium to another.
- Transformation
-
Direct acquisition of DNA from the environment through the cell membrane.
- Gene transfer agents
-
Bacteriophage-like elements that package random DNA regions from a host cell and transfer them to a recipient cell.
- Non-homologous end joining
-
(NHEJ). A pathway for the direct repair of double-strand DNA breaks without a homologous template.
- Nuclear mitochondrial transfers
-
(numts). Transfers of mitochondrial DNA into the nuclear genome of a eukaryotic host; the encoded genes often become non-functional.
- Nuclear plastid transfers
-
(nupts). Transfers of plastid DNA into the nuclear genome of a eukaryotic host; the encoded genes often become non-functional.
- Transposable elements
-
DNA sequences (also known as jumping genes) that can move within a genome (and sometimes also between genomes) by a 'cut and paste' (DNA transposons) or a 'copy and paste' (retrotransposons) mechanism.
- Organellogenesis
-
The process by which an endosymbiont becomes an organelle, in part by becoming (nearly) irreversibly integrated with its host cell at both a genetic and cell biological level.
- Reproductive manipulators
-
Bacteria, such as Wolbachia spp., that are transmitted in the egg cytoplasm of arthropods and nematodes and shift the sex ratio of the host population.
- Peptidoglycan
-
A structural matrix in bacterial cell walls formed by alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) residues, where peptide chains of up to five amino acids link NAM to other NAM-connected peptides.
Rights and permissions
About this article
Cite this article
Husnik, F., McCutcheon, J. Functional horizontal gene transfer from bacteria to eukaryotes. Nat Rev Microbiol 16, 67–79 (2018). https://doi.org/10.1038/nrmicro.2017.137
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2017.137
This article is cited by
-
Salivary proteins potentially derived from horizontal gene transfer are critical for salivary sheath formation and other feeding processes
Communications Biology (2024)
-
Synthetic reversed sequences reveal default genomic states
Nature (2024)
-
Evolutionarily related host and microbial pathways regulate fat desaturation in C. elegans
Nature Communications (2024)
-
Horizontal gene transfer in eukaryotes: aligning theory with data
Nature Reviews Genetics (2024)
-
A mysterious cloak: the peptidoglycan layer of algal and plant plastids
Protoplasma (2024)