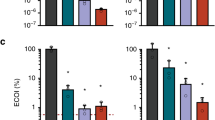
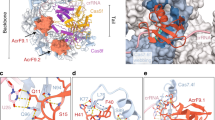
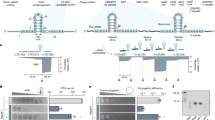
Abstract
CRISPR–Cas adaptive immune systems are widespread among bacteria and archaea. Recent studies have shown that these systems have minimal long-term evolutionary effects in limiting horizontal gene transfer. This suggests that the ability to evade CRISPR–Cas immunity must also be widespread in phages and other mobile genetic elements. In this Progress article, we discuss recent discoveries that highlight how phages inactivate CRISPR–Cas systems by using anti-CRISPR proteins, and we outline evolutionary and biotechnological implications of their activity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Van Valen, L. A new evolutionary law. Evol. Theory 1, 1–30 (1973).
Kruger, D. H. & Bickle, T. A. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol. Rev. 47, 345–360 (1983).
Drozdz, M., Piekarowicz, A., Bujnicki, J. M. & Radlinska, M. Novel non-specific DNA adenine methyltransferases. Nucleic Acids Res. 40, 2119–2130 (2012).
Kahmann, R. The mom gene of bacteriophage Mu. Curr. Top. Microbiol. Immunol. 108, 29–47 (1984).
Studier, F. W. & Movva, N. R. SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J. Virol. 19, 136–145 (1976).
Otsuka, Y. & Yonesaki, T. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 83, 669–681 (2012).
Blower, T. R., Evans, T. J., Przybilski, R., Fineran, P. C. & Salmond, G. P. Viral evasion of a bacterial suicide system by RNA-based molecular mimicry enables infectious altruism. PLoS Genet. 8, e1003023 (2012).
Bondy-Denomy, J., Pawluk, A., Maxwell, K. L. & Davidson, A. R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013).
Pawluk, A. et al. Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829–1838.e9 (2016).
Pawluk, A., Bondy-Denomy, J., Cheung, V. H., Maxwell, K. L. & Davidson, A. R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio 5, e00896 (2014).
Pawluk, A. et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 1, 16085 (2016).
Rauch, B. J. et al. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 168, 150–158.e10 (2017).
Hynes, A. P. et al. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat. Microbiol. http://dx.doi.org/10.1038/s41564-017-0004-7 (2017).
Samson, J. E., Magadan, A. H., Sabri, M. & Moineau, S. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687 (2013).
Garneau, J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Edgar, R. & Qimron, U. The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J. Bacteriol. 192, 6291–6294 (2010).
Brouns, S. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008).
Marraffini, L. A. CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61 (2015).
Mohanraju, P. et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147 (2016).
Wright, A. V., Nunez, J. K. & Doudna, J. A. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell 164, 29–44 (2016).
Makarova, K. S. et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736 (2015).
Koonin, E. V., Makarova, K. S. & Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78 (2017).
Wang, H., La Russa, M. & Qi, L. S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264 (2016).
Bikard, D., Hatoum-Aslan, A., Mucida, D. & Marraffini, L. A. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12, 177–186 (2012).
Wozniak, R. A. & Waldor, M. K. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563 (2010).
Dobrindt, U., Hochhut, B., Hentschel, U. & Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424 (2004).
Juhas, M. et al. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393 (2009).
Sun, C. L. et al. Phage mutations in response to CRISPR diversification in a bacterial population. Environ. Microbiol. 15, 463–470 (2013).
van Houte, S. et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388 (2016).
Fineran, P. C. et al. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl Acad. Sci. USA 111, E1629–E1638 (2014).
Gophna, U. et al. No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. ISME J. 9, 2021–2027 (2015).
Touchon, M., Bernheim, A. & Rocha, E. P. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 10, 2744–2754 (2016).
Touchon, M. et al. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology 158, 2997–3004 (2012).
Dang, T. N. et al. Uropathogenic Escherichia coli are less likely than paired fecal E. coli to have CRISPR loci. Infect. Genet. Evol. 19, 212–218 (2013).
Bondy-Denomy, J. et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 (2015).
Chowdhury, S. et al. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell 169, 47–57.e11 (2017).
Peng, R. et al. Alternate binding modes of anti-CRISPR viral suppressors AcrF1/2 to Csy surveillance complex revealed by cryo-EM structures. Cell Res. 27, 853–864 (2017).
Maxwell, K. L. et al. The solution structure of an anti-CRISPR protein. Nat. Commun. 7, 13134 (2016).
van Erp, P. B. et al. Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res. 43, 8381–8391 (2015).
Dong, D. et al. Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature 546, 436–439 (2017).
Yang, H. & Patel, D. J. Inhibition mechanism of an anti-CRISPR suppressor AcrIIA4 targeting SpyCas9. Mol. Cell 67, 117–127.e5 (2017).
Shin, J. et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 3, e1701620 (2017).
Harrington, L. B. et al. A broad-spectrum inhibitor of CRISPR-Cas9. Cell 170, 1224–1233.e15 (2017).
Wang, X. et al. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat. Struct. Mol. Biol. 23, 868–870 (2016).
Wang, J. et al. A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses. Cell Res. 26, 1165–1168 (2016).
Burstein, D. et al. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat. Commun. 7, 10613 (2016).
Jiang, W. et al. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 9, e1003844 (2013).
Westra, E. R. et al. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr. Biol. 25, 1043–1049 (2015).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Dominguez, A. A., Lim, W. A. & Qi, L. S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 17, 5–15 (2016).
Maxwell, K. L. Phages fight back: inactivation of the CRISPR-Cas bacterial immune system by anti-CRISPR proteins. PLoS Pathog. 12, e1005282 (2016).
Hug, L. A. et al. A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016).
Acknowledgements
A.P. was supported by a Canadian Institutes of Health Research Doctoral Award. Research in this area in the authors' laboratories is supported by Canadian Institutes of Health Research grants to A.R.D. (MOP-130482) and K.L.M. (MOP-136845).
Author information
Authors and Affiliations
Contributions
A.P. researched the data for the article. A.P. and K.L.M. wrote the article. A.P., K.L.M. and A.R.D. substantially contributed to discussions of the content and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare potential competing interest as all authors have a patent pending for anti-CRISPR technologies on which they are inventors.
Rights and permissions
About this article
Cite this article
Pawluk, A., Davidson, A. & Maxwell, K. Anti-CRISPR: discovery, mechanism and function. Nat Rev Microbiol 16, 12–17 (2018). https://doi.org/10.1038/nrmicro.2017.120
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2017.120
This article is cited by
-
Alternative therapeutic strategies to treat antibiotic-resistant pathogens
Nature Reviews Microbiology (2024)
-
An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding
Nature Communications (2024)
-
Insights into the inhibition of protospacer integration via direct interaction between Cas2 and AcrVA5
Nature Communications (2024)
-
Development of Cas13a-based therapy for cancer treatment
Molecular Biology Reports (2024)
-
The highly diverse antiphage defence systems of bacteria
Nature Reviews Microbiology (2023)